Digital PCR is a specialized approach to nucleic acid detection and quantification that estimates absolute numbers of molecules through statistical methods. Digital PCR (dPCR) uses the same fundamental chemistry as qPCR, but unlike qPCR data, dPCR data are collected at the endpoint of the reaction mix. Before amplification, a bulk PCR reaction made up of nucleic acid, primers, probes, and master mix is digitized into many thousands of nanoliter-sized microreactions. As this digitization process distributes the PCR mix across so many microreactions, each microreaction will effectively either contain one, zero, or just a handful of the target nucleic acid molecules. The isolated microreactions are then amplified, and data are collected from each microreaction at the end of the thermal cycling process. Microreactions that do not contain the target will not show post-amplification fluorescence, while those that do contain the target will show post-amplification fluorescence.
In effect, the original reaction is turned into many binary reactions. After counting the positive microreactions, simple statistics can be used to then determine the “absolute” quantity of the target molecule rather than a quantity estimated by comparing to a standard of known concentration. This technology offers an alternative to qPCR for absolute quantification and rare allele detection which does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample.
To date, qPCR has been a powerful and sensitive gene analysis technique used for a broad range of applications. As the name suggests, qPCR measures PCR amplification as it occurs, unlike traditional PCR, which collects results after the reaction is complete, making it impossible to determine the starting concentration of nucleic acid. qPCR’s entry into the market revolutionized PCR-based quantitation of DNA and RNA.
Digital PCR vs. Real-Time PCR vs. Traditional PCR
To date, qPCR has been a powerful and sensitive gene analysis techniques used for a broad range of applications. As the name suggests, qPCR measures PCR amplification as it occurs, unlike traditional PCR, which collects results after the reaction is complete, making it impossible to determine the starting concentration of nucleic acid. qPCR’s entry into the market revolutionized PCR-based quantitation of DNA and RNA.
Digital PCR is a newer approach to nucleic acid detection and quantification that estimates absolute numbers of molecules through statistical methods. This technology offers an alternative to qPCR for absolute quantification and rare allele detection rather than relying on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample.
|
Digital PCR vs. Real-Time qPCR vs. Traditional PCR at a Glance |
|||
|
Digital PCR |
Real-Time qPCR |
Traditional PCR |
|
| Overview |
Measures the fraction of negative microreactions to determine absolute copies. |
Measures PCR amplification as it occurs in a bulk reaction mix. |
Measures the amount of accumulated PCR product at the end of the PCR cycles. |
| Quantitative? |
Yes – The fraction of negative microreactions is fit to a Poisson statistical algorithm. |
Yes – Data is collected during the exponential growth phase of PCR when the quantity of the PCR product is directly proportional to the amount of template nucleic acid. |
No – However, comparing the intensity of the amplified band on a gel to standards of a known concentration can give you semi-quantitative results. |
| Applications |
|
|
|
| Advantages |
|
|
|
How Digital PCR Works
dPCR Counts Individual Molecules for Absolute Quantification
Whereas traditional PCR measures at the plateau, giving you variable results due to variations in reaction kinetics, and qPCR measures at the exponential phase for more accurate quantitation, dPCR counts individual molecules for absolute quantification.
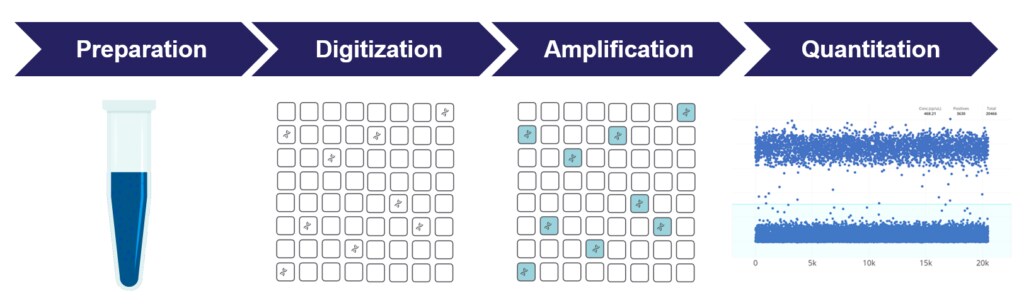
dPCR and the Importance of Individual Microreactions
dPCR works by dividing a sample of DNA, cDNA, or RNA into many individual microreactions; some microreactions contain one or more molecules while others contain none. Each microreaction undergoes PCR amplification and analysis separately. Microreactions with and without amplified product are individually counted. Those containing amplified product are designated positive; those with no amplified product are designated negative.
Microfluidic Plate to Run Thousands of Microreactions in Parallel
A microfluidic plate provides a convenient and straightforward mechanism to run thousands of microreactions in parallel. Each upstream well of the plate is loaded with a mixture of sample, master mix, and reagents, and after distribution into microreactions, individually analyzed to detect the presence or absence of an endpoint signal.
Following PCR analysis, the fraction of negative microreactions is used to generate an absolute quantity of the number of target molecules in the sample, without standards or endogenous controls.
Correction Factor to Determine Absolute Template Quantity
Poisson Model
Due to random assortment, there’s no assurance that each positive reaction received only a single molecule. To account for microreactions that may have received more than one molecule of the target sequence, a correction factor to determine the absolute template quantity is applied using the Poisson model.
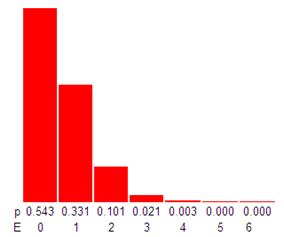
In the case of the Applied Biosystems™ QuantStudio™ Absolute Q™ Digital PCR system, each reaction of the Applied Biosystems™ QuantStudio MAP16 dPCR plate contains 20,480 fixed microchambers. After performing the dPCR reaction, the QuantStudio Absolute Q Digital PCR Analysis software sets the thresholds and performs the calculation.

Using the Poisson Model
To apply the Poisson model, you must have at least one negative reaction – a reaction with no molecule. The Poisson model describes the probability of a reaction receiving zero, one, two, or three copies (see bar chart above). The model corrects for reactions containing multiple molecules and provides a probability that the answer is correct. As a result, sample digitization paired with Poisson statistical data analysis allows higher precision than traditional PCR and qPCR methods.
It’s important to note that when using the Poisson model, the negative reactions matter most (see 96 well plate results above). They help establish both the number of DNA molecules present in the original sample but also the ratio of positive to negative reactions post PCR analysis.
For example, dPCR is particularly well suited for applications that require the detection of small amounts of input nucleic acid or finer resolution of target amounts among samples, eg, for cancer research applications rare sequence detection of cancer mutations present at very low quantities in liquid biopsy samples.
Related: Mutation Detection Using Digital PCR
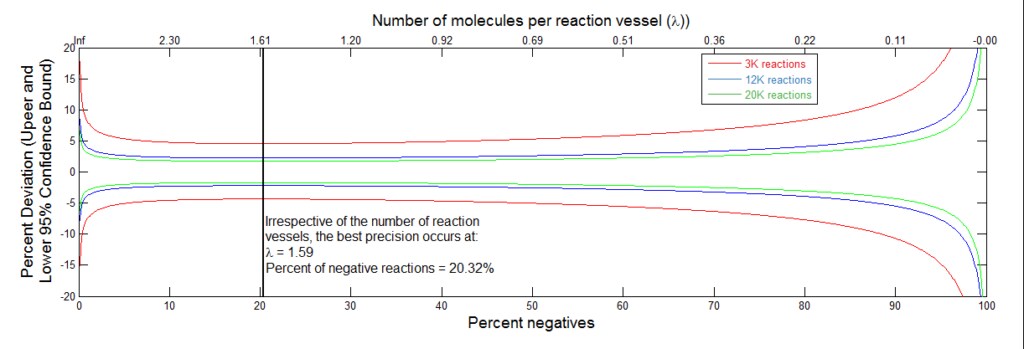
CAPTION: For any digital PCR reaction you have maximum precision when you look at 1.6 copies per reaction. The graph shows precision that one would achieve using either 20K (green), 12K (blue) or 3K (red) microreactions per sample. The X-axis represents negative reactions (20.32%) while the Y-axis represents precision (95% confidence). Maximum precision comes out to 20% negatives and 80% positives. Looking at the green line, precision is consistent between ~5% and 90% negative reactions 20K microreaction per sample. To increase the number of microreactions per sample, digital pooling may be leveraged to improve precision. This is the target to aim for with your sample concentration, e.g., one copy per reaction well – 20,000 reactions in 15 uL. At either extreme, precision drops off precipitously.
Digital PCR Delivers High Levels of Accuracy and Reproducibility
Before we talk about applications, here’s data that demonstrates digital PCR’s reproducibility.
The graph below shows the reproducibility of dPCR in absolute quantification. The graph shows the results of an absolute quantification experiment run on 24 plates across 3 different users. As you can see that these three different users were able to obtain basically the same results.

Digital PCR Works With a Range of Applications
dPCR is well suited to performing rare allele detection, measurement of copy number variation, viral titer measurement, quantification of next-generation sequencing libraries, and detecting rare targets from environmental samples such as wastewater.
Specific applications available for absolute quantification include:
- Copy Number Variation (CNV) – Detect and quantify small percent copy number differences with a high degree of precision
- Next Generation Sequencing (NGS) Library Quantification – Absolute quantification of NGS libraries and validation of sequencing results, without reference standards
- Rare target quantification – Detect and quantify rare mutations for low-prevalence targets in cancer research samples
- Quantification of Viral Titer – Absolute quantification of viral vector backbones commonly used in cell and gene therapy research such as adenoassociated virus (AAV) and cytomegalovirus (CMV)
- Environmental Pathogen Detection – Detect pathogenic viruses and bacteria from environmental samples such as wastewater or sewage samples
Summary of dPCR Advantages
dPCR Provides a Mechanism for Absolute Quantification
First and foremost, dPCR provides a mechanism for absolute quantification, which lets you determine the number of molecules in your sample without the need for standard curve. This is especially beneficial for reference laboratories and gene expression analysis.
dPCR Allows Rare Molecule Identification
Secondly, dPCR allows you to identify a rare molecule in an overwhelming number of normal alleles such as detecting oncogenic mutations in circulating free DNA.
dPCR is Less Sensitive to Inhibitors
Finally, because dPCR is an endpoint PCR reaction, it’s less sensitive to inhibitors than qPCR reactions, where inhibitors such as SDS or heparin could affect PCR efficiency. This is an important attribute for research labs working with environmental or precious cancer samples.
Learn More About Digital PCR
- Download Digital PCR Technical Literature
- Compare and contrast Digital PCR vs. qPCR vs. Traditional PCR – Three generations of PCR technology.
- Get info on QuantStudio Absolute Q Digital PCR System – Learn about this powerfully simple digital PCR system. This single instrument system has a simple qPCR-like workflow, fast 90-minute time to answer, and reference curve-free quantitation providing concentration measurements in copies/μL without a standard curve.
- Watch this short video on Next-generation Digital PCR Workflow – See for yourself how fast and simple digital PCR can be using the QuantStudio Absolute Q Digital PCR System.
- Learn more about Powerfully Simple dPCR for Absolute Quantification – Complete information about digital PCR and relevant applications, including new and relevant technical and applications notes, videos, and resources.



Leave a Reply