Search
Excerpts from RNA Interference Research Guide
Much of this article was taken from Ambion's newly available RNA Interference Research Guide, which provides background information on RNAi, guidelines for optimizing siRNA delivery, recommendations for controlling siRNA experiments, an overview of siRNA screening, and detailed descriptions of the necessary tools required from start to finish for successful RNAi experiments. Request a free RNA Interference Research Guide today.
RNA interference, the biological mechanism by which double-stranded RNA (dsRNA) induces gene silencing by targeting complementary mRNA for degradation, is revolutionizing the way researchers study gene function. For the first time, scientists can quickly and easily reduce the expression of a particular gene in nearly all metazoan systems, often by 90% or greater, to analyze the effect that gene has on cellular function. The ease of the technique, as well as the wide availability of high quality kits and reagents for performing RNAi experiments, has driven its incredibly rapid adoption by the research community.
Reagents needed for RNAi experiments
- A dsRNA (i.e., siRNA or long dsRNA) that is completely complementary to the gene transcript(s) you wish to target by RNAi
- A means to deliver that dsRNA to cells
- Proper controls
- A way to detect the biological effect of reducing target gene expression (i.e., an assay).
siRNA design and synthesis for mammalian RNAi experiments
For simplicity, we will confine our discussion to short term experiments in mammalian cell lines where RNAi is typically induced using short interfering RNAs (siRNAs) complementary to a desired target mRNA (Figure 1; for a more complete discussion of the mechanism and history of RNAi, see www.ambion.com/RNAi). siRNAs are generally 21 bp double-stranded RNA molecules with dinucleotide 3' overhangs, and are generated intracellularly in the RNAi pathway when the nuclease Dicer cleaves long dsRNA. In mammalian cells, siRNAs have been elevated from their usual role as an RNAi intermediary to become the primary RNAi trigger used by researchers, since the long dsRNA that successfully induces RNAi in Caenorhabditis elegans and Drosophila induces a potent antiviral response in mammalian cells. siRNAs used to induce RNAi in mammalian systems are most often synthesized chemically by RNA oligonucleotide manufacturers such as Ambion, however they can also be expressed as short hairpin RNAs from a DNA construct.
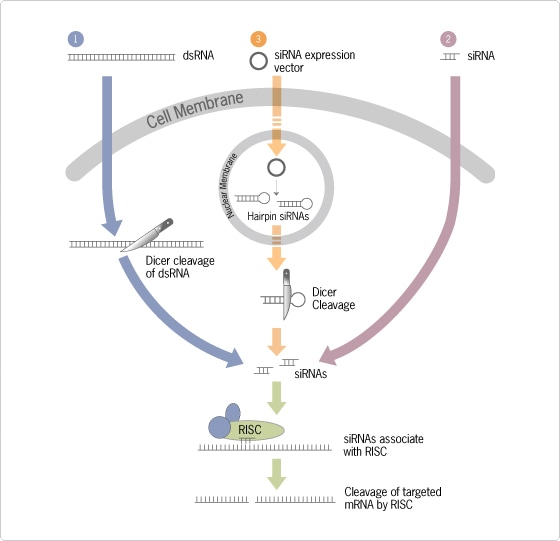
Figure 1. Three Ways to Trigger the RNAi Pathway. (1) In non-mammalian systems, the RNAi pathway commences when double-stranded RNA (dsRNA; usually longer than 30 bp) is introduced into cells. In mammalian systems, RNAi can be triggered by synthetic short interfering RNA (siRNA) molecules (2) or by DNA based expression vectors designed to express short hairpin RNA (shRNA) molecules (3). In each case, gene silencing results from destruction of mRNA that is complementary to the input siRNA (2) or the siRNA molecules created by Dicer cleavage of longer dsRNA (1) or shRNA (3) molecules. See text for additional details. Dicer=cytoplasmic nuclease; RISC=RNA-induced silencing complex; mRNA=messenger RNA.
A few companies have developed complex "intelligent algorithms" that have proven to be effective at designing efficacious siRNAs. Ambion uses one of the most broadly validated intelligent algorithms to provide guaranteed-to-silence siRNAs for all human, mouse, and rat genes (see [1] for more details about this algorithm). These ready-to-use, chemically synthesized siRNAs are available individually as Silencer Pre-designed siRNAs or Silencer Validated siRNAs, and in genome-wide, functional class-focused, and custom sets as Silencer siRNA Libraries and Silencer CellReady siRNA Libraries. Individual siRNAs allow detailed analysis of an individual gene's role in one or more pathways, whereas siRNA libraries, or sets of siRNAs targeting a pre-defined or custom set of genes, enable large scale screening experiments to correlate genes with cellular function.
Delivery of siRNAs
Once you have an siRNA, you need a means to deliver it, and the siRNA delivery conditions need to be tested and often optimized for each cell type being used. siRNAs can be introduced directly into cells by transfection or electroporation. For many immortalized cell lines, transfection with a lipid- or amine-based reagent is the preferred option. Delivery into primary cells and suspension cells, however, often requires electroporation using a specialized, gentle-on-cells buffer, such as siPORT siRNA Electroporation Buffer, and optimized pulsing conditions. Such conditions generally result in very efficient siRNA delivery without compromising cell viability.
Controls for siRNA experiments
Assay for RNAi effect
RNAi resources
For Research Use Only. Not for use in diagnostic procedures.