Search
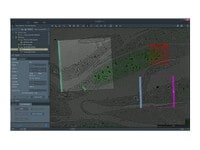
El conocimiento de la compleja arquitectura 3D de células y tejidos en su contexto nativo es esencial para comprender las relaciones biológicas entre la estructura y la función. Thermo Fisher Scientific ofrece una novedosa solución de adquisición de imágenes de cara de bloques en serie (SBFI) que combina la microscopía electrónica de barrido de deconvolución de la multienergía (MED-SEM) con cortes en secciones in situ. La automatización y la facilidad de uso aumentan su productividad, independientemente de su nivel de experiencia, lo que le otorga una resolución isotrópica para sus muestras de gran volumen.
Hasta ahora, la resolución axial en las adquisición de imágenes de cara de bloques en serie estaba limitada por el grosor mínimo de la sección que se puede cortar físicamente con un micrótomo en la cámara. Sin embargo, gracias a la incorporación de la deconvolución de la multienergía SEM (MED-SEM), nuestro novedoso método de análisis de grandes volúmenes permite ahora la adquisición de imágenes con una resolución 3D verdaderamente isotrópica. El corte en secciones mecánico y óptico se combinan; los datos se adquieren a partir de múltiples capas de la muestra mediante la aplicación de diferentes energías de haz entre cortes físicos. Los detectores optimizados y el funcionamiento con vacío bajo garantizan una adquisición de imágenes de alta calidad. Además, el software de adquisición proporciona automatización de procesos que van desde la configuración y alineación de bajo nivel hasta la adquisición de una serie de imágenes completa. Este enfoque incluye un flujo de trabajo completo desde la configuración inicial hasta los resultados finales, proporcionando soluciones de software para análisis de grandes áreas/volúmenes, superposición de imágenes de microscopía óptica, reconstrucción, visualización y segmentación.


Retina de ratón proyectada con análisis de grandes volúmenes. Dimensiones: 9,65 x 10 x 25 µm; resolución isotrópica de 10 nm en modo HiVac; 1,18/1,78/2,27 kV, 100 pA; tiempo de radiación de 1 µs; 250 imágenes.
Cabeza de embrión de pez cebra con imágenes de grandes volúmenes. Dimensiones: 350 x 350 x 82,9 µm. 42 x 42 nm píxeles en modo LoVac, 2 kV, 100 pA, 3 µs de tiempo de permanencia, 829 imágenes a 100 nm. Muestra cortesía de Robbert Creton, Universidad Brown.
Cerebro de rata: Volumen 85 x 85 x 123 um; 2,7 kV, 400 pA, 2 us de tiempo de permanencia, 15 nm x 15 nm x 40 nm, 2133 imágenes en LoVac. Muestra cortesía de Grahame Knott, EPFL Lausanne. Segmentación y visualización de datos con el software Amira de Thermo Scientific.
Funcionamiento
Descargas
Retina de ratón proyectada con análisis de grandes volúmenes. Dimensiones: 9,65 x 10 x 25 µm; resolución isotrópica de 10 nm en modo HiVac; 1,18/1,78/2,27 kV, 100 pA; tiempo de radiación de 1 µs; 250 imágenes.
Cabeza de embrión de pez cebra con imágenes de grandes volúmenes. Dimensiones: 350 x 350 x 82,9 µm. 42 x 42 nm píxeles en modo LoVac, 2 kV, 100 pA, 3 µs de tiempo de permanencia, 829 imágenes a 100 nm. Muestra cortesía de Robbert Creton, Universidad Brown.
Cerebro de rata: Volumen 85 x 85 x 123 um; 2,7 kV, 400 pA, 2 us de tiempo de permanencia, 15 nm x 15 nm x 40 nm, 2133 imágenes en LoVac. Muestra cortesía de Grahame Knott, EPFL Lausanne. Segmentación y visualización de datos con el software Amira de Thermo Scientific.
Funcionamiento
Descargas
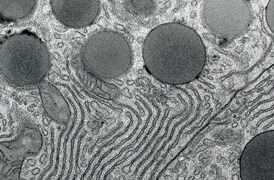
Investigación sobre patologías
La microscopía electrónica de transmisión (TEM) se utiliza cuando la naturaleza de la enfermedad no puede establecerse mediante métodos alternativos. Con las imágenes nano-biológicas, TEM proporciona información precisa y fiable para ciertas patologías.

Investigación sobre biología de plantas
La investigación fundamental de la biología de las plantas se hace posible mediante la criomicroscopía electrónica, que proporciona información sobre proteínas (con análisis de partículas individuales), a su contexto celular (con tomografía), hasta la estructura general de la planta (análisis de grandes volúmenes).