Search
Products for RNA Structure/Function Analysis
RNA Structure Has a Biological Role
RNA structure is thought to play a central role in many cellular processes, including transcription initiation, elongation and termination, mRNA splicing, and retroviral infection of eukaryotic cells. Perhaps the most striking example of RNA structure being critical to an essential cellular process is the ribonucleoprotein complex that constitutes the translational machinery. Ribosomal RNA (rRNA) molecules act as a scaffold on which proteins are assembled to form a functional ribosome. There is mounting evidence that rRNA forms the catalytic center of the peptidyl transferase activity. In the last 5 years, RNA molecules have been found to be crucial in newly discovered biological phenomenon in unexpected and fascinating ways. Some of these include: RNA interference (RNAi), in which genes are silenced post-transcriptionally in plants and animals by double-stranded RNA (dsRNA) that is homologous in sequence; long-distance plant signal trafficking, involving plant phloem packed with RNA molecules; and RNA transport where the RNA transcript is assembled into a ribonucleoprotein fibril, compacted into a ring-like structure, that gradually unfolds through the nuclear pore to be finally recruited for protein synthesis.
RNA structure is thought to play a central role in many cellular processes, including transcription initiation, elongation and termination, mRNA splicing, and retroviral infection of eukaryotic cells. Perhaps the most striking example of RNA structure being critical to an essential cellular process is the ribonucleoprotein complex that constitutes the translational machinery. Ribosomal RNA (rRNA) molecules act as a scaffold on which proteins are assembled to form a functional ribosome. There is mounting evidence that rRNA forms the catalytic center of the peptidyl transferase activity. In the last 5 years, RNA molecules have been found to be crucial in newly discovered biological phenomenon in unexpected and fascinating ways. Some of these include: RNA interference (RNAi), in which genes are silenced post-transcriptionally in plants and animals by double-stranded RNA (dsRNA) that is homologous in sequence; long-distance plant signal trafficking, involving plant phloem packed with RNA molecules; and RNA transport where the RNA transcript is assembled into a ribonucleoprotein fibril, compacted into a ring-like structure, that gradually unfolds through the nuclear pore to be finally recruited for protein synthesis.
Structural Characterization of RNA
Elucidating the mechanistic aspects of these intricate processes will require detailed understanding of the underlying RNA structure. RNA molecules usually possess a variety of single-stranded and double-stranded regions that give rise to complex three-dimensional structures. These structures probably define the molecule's interactions with other nucleic acids, proteins, and small molecules.
The types of experiments performed to structurally characterize RNA utilize a variety of reagents that are neither available from a single source nor are optimized for RNA applications. Ambion is committed to providing high quality kits and reagents for these applications. We also hope to spark interest among those unaware of the techniques used for RNA characterization.
The types of experiments performed to structurally characterize RNA utilize a variety of reagents that are neither available from a single source nor are optimized for RNA applications. Ambion is committed to providing high quality kits and reagents for these applications. We also hope to spark interest among those unaware of the techniques used for RNA characterization.
Biochemistry-Grade Ribonucleases
- Defining RNA structure
- Sequencing RNA
- RNA:Protein Footprinting
- Boundary experiments
Ambion now offers RNases A, V1, and T1 that have been optimized for researchers performing RNA structural analysis, RNA sequencing, protein footprinting and boundary experiments. RNase A cleaves 3' of single-stranded C and U residues. RNase V1 cleaves base-paired nucleotides. RNase T1 cleaves 3' of single-stranded G residues. Combinations of the single-strand and double-strand specific ribonucleases can provide rapid analysis of the physical structure of an RNA of interest (Figure 1). These ribonucleases can also be used to map protein-binding sites on RNAs by comparing cleavage patterns in the presence and absence of an RNA binding protein. RNase T1 by itself can be used to define the boundaries of the minimal RNA sequence required for selectable activities such as protein binding or catalysis.
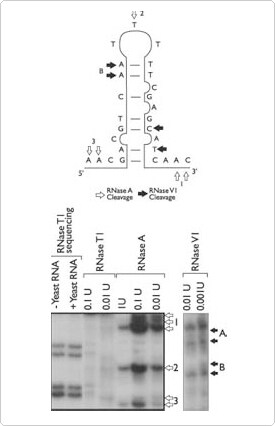
Figure 1. Structural Analysis of an RNA. The RNA molecule shown above was end-labeled and subjected to digestion with RNases A, TI, and VI. The resulting fragments were visualized by denaturing PAGE.
Modified Nucleotides
Modified nucleotides that confer unique characteristics on the RNA molecules into which they are incorporated can be extremely useful tools for biochemical and molecular biology studies. Modified RNA transcripts have been used to study RNA catalysis, protein binding and RNAi. Ambion now offers a variety of modified NTPs that can confer nuclease stability, chemical instability, and enhanced crosslinking or hybridization properties on the transcript into which they are incorporated.