
The importance of fragmentation in mass spectrometry
1. What is fragmentation in the context of mass spectrometry?
Fragmentation in this sense is the process of breaking a molecule into smaller pieces inside a mass spectrometer. By applying energy to break bonds, we create fragments that provide critical clues about the molecule’s structure, helping scientists piece together the complete picture of an unknown substance.
These fragments form a spectrum, a unique fingerprint of the molecule, which helps identify its components, modifications, and overall structure.
2. Why does fragmentation matter?
Without fragmentation, you would only know the intact mass of a molecule. This can provide some information, but for identifying something as complex as a peptide, it would be challenging to know the exact amino acid sequence. In a standard tryptic peptide search, using a HeLa library, there are up to 20 peptides overlapping within each 10 ppm error bin. (FIGURE 1) Fragmentation becomes essential for getting the full story since the peptides within a reasonable mass error may elute at the same time, or have less predictable elution, so the matching by MS1 only yields too many results.
This does not even account for the addition of modifications into the search space. Just to give context, there are a huge number of putative peptides with modifications1 (Figure 2), many of which would benefit from the use of alternate fragmentation techniques to help identify and characterize. We will discuss many of these in more detail in other sections. Without fragmentation, there would be no way to distinguish many peptides from one another to make an identification with confidence.
Think of a peptide like a radio. The intact mass is the cover – it tells you it’s a radio, but it doesn’t show you how it works or what’s inside. Without taking the radio apart, you could put the same cover on a cake and think you might have two identical radios! To truly understand it and build a replica, you have to take it apart, piece by piece. That’s exactly what fragmentation does in mass spectrometry.
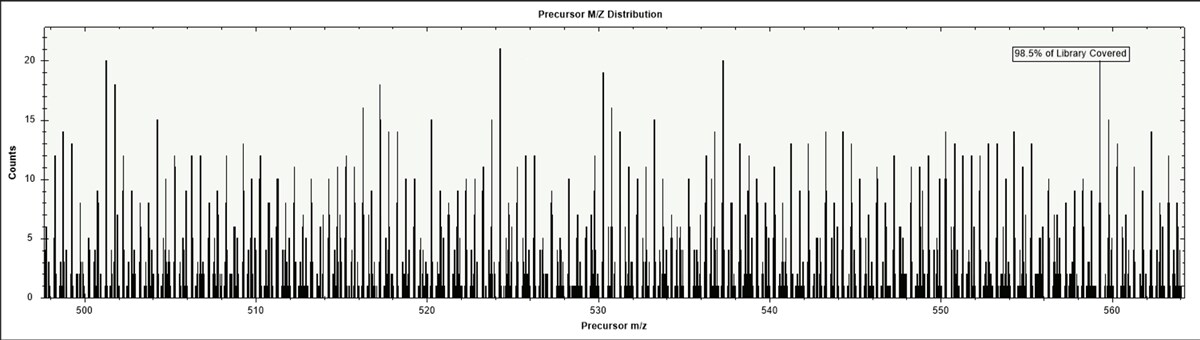
Figure 1: HeLa library of tryptic digested peptides binned by 10 ppm error, highlighted here is the 500-550 m/z range, showing individual bins with up to 20 potential peptide sequence matches.
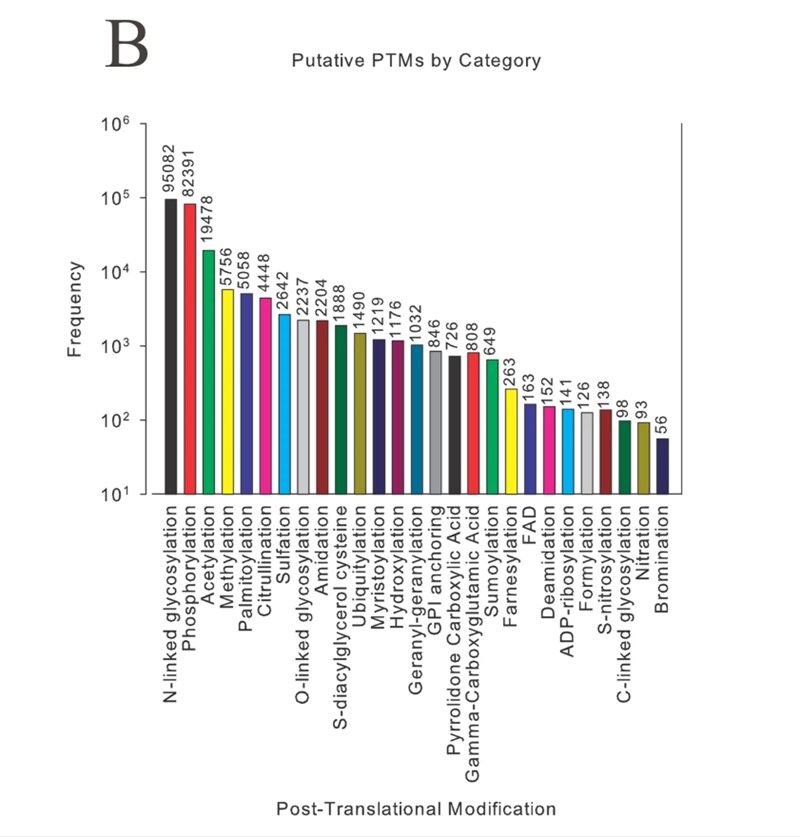
Figure 2: Putative post-translational modifications (PTMs) from the Swiss-Prot database listed in descending order.1
3. What applications rely on alternative or multiple fragmentation types?
Fragmentation is a well-established aspect of mass spectrometry, but some systems provide many options for fragmentation, while others may only have one or even no options for fragmentation. The need within this space all depends on the application being studied.
Most proteomics experiments get rich, meaningful fragments from quick, efficient, and common collision-based methods. On the other hand, proteomics samples rich in glycosylation will break too easily, destroying the ability to map back the modification, and so more gentle techniques, like electron-based ones, need to be employed.
Generally speaking, when scientists need a better characterization of their analyte, they rely on multiple or alternate fragmentation techniques to do so. Glycomics, glycoproteomics, lipidomics, top-down proteomics, and metabolomics commonly rely on alternate or complementary fragmentation for complete characterization of the molecules.
One of the most common fragmentation applications is on proteins and peptides. They fragment predictably along their backbone to create a, b, c, or x, y, z fragment ions. [Figure 3]. A, b, and c type fragments are those that include from the n-terminus of the peptide to the amino acid residue number, reading from left to right. X, y, and z type fragments are those that include the c-terminus of the peptide to the amino acid residue number, reading from right to left.
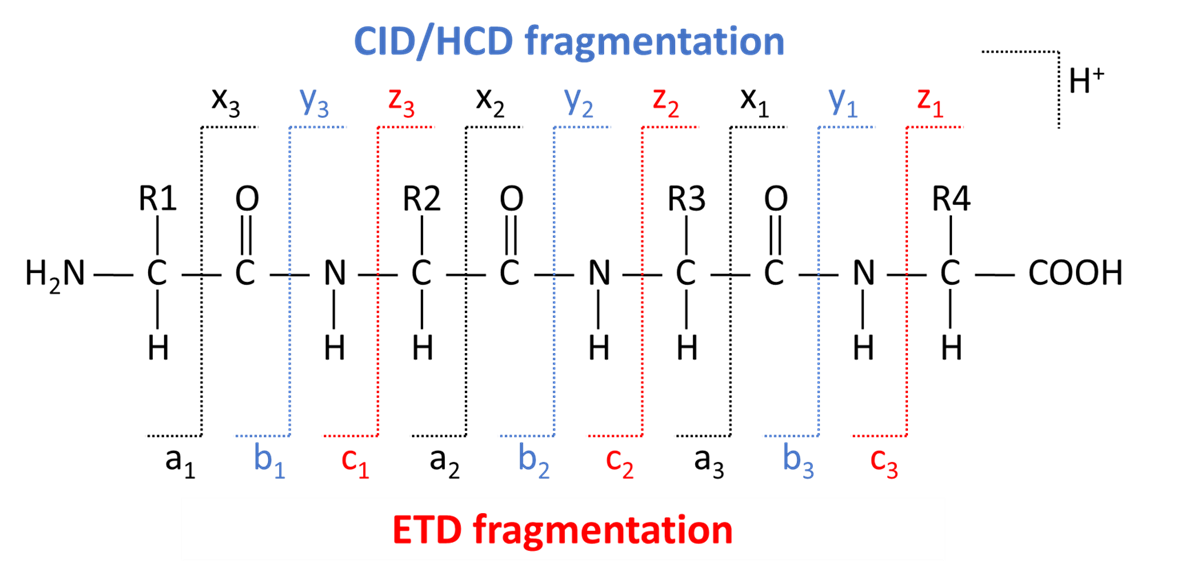
Figure 3: Location of peptide/protein backbone cleavage and its nomenclature. Representative here is a 4-amino acid peptide with side chains labeled R1-4.
In this series, we will cover the basics, principles, applications, and considerations for the alternate fragmentation and ion-manipulation techniques available on Thermo Scientific mass spectrometers. Up next, collision-based fragmentation.
Learn more about Freedom to Fragment
References:
Visit us on LinkedIn: #Fragmentation #ETD



