This is the second part of a multi-part Library blog series. For additional information, see blog Part 1- Generating Quantitative Methods Automatically From Spectral Library Data and stay tuned for more blogs on our website.
While identification of unknowns remains a significant challenge, it has become easier over the years with the growth of large MSn spectral libraries. These libraries, such as mzCloud, can provide substructure data on unknowns. Such information can help us to elucidate the structure of compounds that would previously remain unknown.
Substructure identification such as this takes advantage of the fact that a structure will always create the same fragments under a given set of fragmentation conditions regardless of if that structure was from MS1 or an MSn level of fragmentation. While no library will ever contain fragmentation data on all possible chemicals, what we need with this approach is enough breadth of coverage in the various substructures commonly occurring to “build up” larger molecules. What that means is we can take the MSn data on many structurally diverse molecules and use it to identify substructures in our unknown.
As an example of using MSn data in this fashion, figure 1 shows the query MSn spectra for an unknown and two of the library hits. The hits are from MSn spectra of two known compounds and the precursor of the specific MSn fragment spectra is shown. Both hits from the library provide a similar structure – an aromatic benzyne linked to either an amide or linked to a carbonyl and amine group.
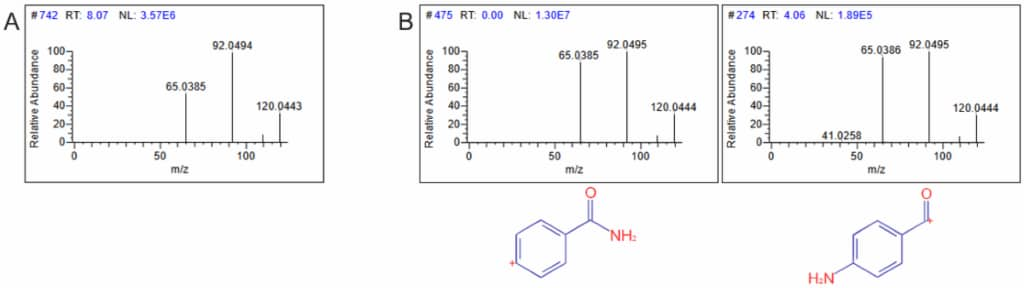
Figure 1: The MS3 spectra of the unknown (A) and two matching fragment spectra from the library search along with the precursors of those spectra (B).
While in this example, the specific orientation could not be determined, we have at least a very good starting point in our structure determination by knowing at least one piece of our unknown. Further searches with other MSn spectra from our molecule may uncover other substructures bringing us close to a final identification. In fact, the unknown in this case was isoproturon (Figure 2) which indeed has the substructure indicated from the search.
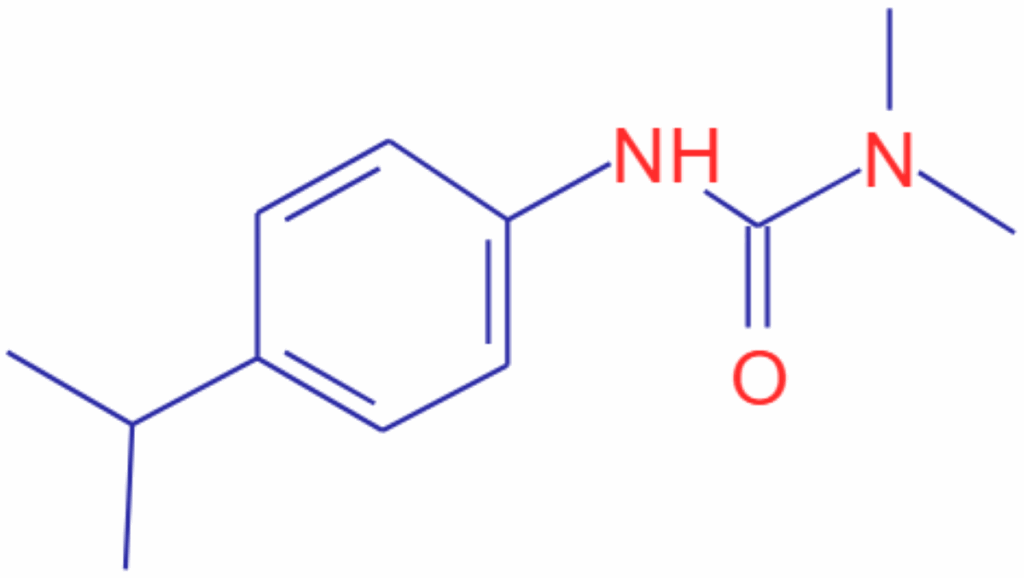
Figure 2: Structure of Isoproturon
By utilizing a large library of MSn reference data and acquiring the same MSn query data on our unknowns, we can begin to build up a picture based on substructures we identify. Visit mzCloud to see the breadth of MSn fragmentation spectra on a diverse range of chemicals that can be used for such an approach.
Visit us on LinkedIn: #mzCloud #MassSpectralLibraries