
The Ion GeneStudio S5 Next-Generation Sequencing Systems allow clinical researchers the ability to show their versatility in various scientific undertakings. Next-Generation Sequencing (NGS) libraries are clonally templated and loaded onto a series of semi-conductor chips using the Ion Chef System. A prepared semi-conductor chip can now be sequenced by The Ion GeneStudio S5 Next-Generation Sequencing Systems. The detection of the removal of the 3’-hydroxyl group during nucleic base addition without fluorescence is the basis for the technology
The group at University Hospitals Translational Laboratory (UHTL) in Cleveland, Ohio is using the flexibility of the Ion GeneStudio S5 Prime System to conduct research that investigate retrovirus infections in the real world. They are using the Ion 510 chip, the smallest-throughput chip of the Ion sequencing chip series. The Ion 510 chip can generate 2–3 million reads of sequencing data, with read lengths up to 400 bp and rapid sequencing run times of 2.5-4 hours. With this capability UHTL conducts their HIV genotyping assay that determines HIV-1 susceptibility for antiretroviral drugs targeting protease, reverse transcriptase, integrase and maturation inhibitors, as well as predicting HIV-1 co-receptor tropism.

Figure 1: Five different sequencing chips enable a throughput range from 2M to 130M reads per run
Additionally, UHTL has shown that the Ion 510 chip produces reliable sequencing data that allows researchers to make stomach disease assessments.
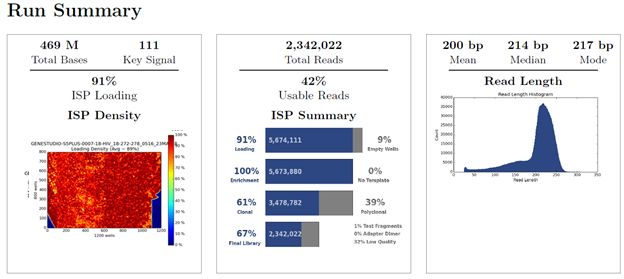
Figure 2: Ion 510 chip run data for HIV Genotyping Assay showing 2.3M READS on Ion GeneStudio S5 Prime System and Ion Chef System
Chronic bacterial infections with Helicobacter Pylori (H. Pylori) is the main etiology of peptic ulcer disease, the most common stomach disease, and the trigger for malignant diseases such as gastric MALT lymphoma. Their custom designed NGS assay detects the presence of H. pylori and mutations in H. Pylori genome that confer antibiotic resistance to commonly used antibiotics using FFPE gastric biopsy samples. This assay is performed by multiplexing 15-20 samples on a single Ion 510 chip.
For Research Use Only. Not for use in diagnostic procedures.



