
Effective DNA extraction is the cornerstone of molecular biology research, diagnostics, and forensic applications worldwide. Whether you’re working with blood samples in a clinical lab, plant tissues in agricultural research, or FFPE samples in cancer studies, each sample type presents unique challenges that demand specialized approaches.
In this guide, we will explore the importance of DNA purification and highlight the advanced solutions to meet the diverse needs of researchers in the biotech field.
Table of contents
Why is DNA extraction quality important?
DNA analysis is a keystone for a huge range of scientific applications, including:
- Exploration of genetic research with human genome mapping, gene cloning, and functional genomics
- Development of future clinical diagnostics with discovery in disease research, pathogen discovery, development of personalized medicine
- Forensic analysis with DNA profiling and paternity testing
- Pharmaceutical development across gene therapy and recombinant protein production
- Development of agricultural biotechnologies in genetic engineering and marker-assisted selection
- Exploration of environmental biotechnologies with biodiversity studies and bioremediation
Without quality extraction, none of these applications would be possible.
DNA extraction process at-a-glance
There are several ways to isolate DNA including organic isolation, isolation by filtration or precipitation, and isolation with magnetic bead sources. All follow a standard order of operations:
- Cellular lysis
- Binding or precipitation
- Washing of bound or precipitated DNA
- Elution or resuspension into working buffer for downstream use
DNA extraction challenges by sample type
Different sample types present specific challenges for DNA isolation, compounded by variables such as sample volume, DNA size, and required target concentration for downstream analysis.
For example, blood samples contain inhibitors that can affect downstream processes. Plant tissues have rigid cell walls that complicate cell lysis. Tissue samples often suffer from DNA degradation, and microbial samples feature robust cell walls that resist standard extraction methods.
Each sample type—blood, tissue, cell cultures, plant material, and microbial samples—requires specialized approaches to ensure efficient, high-quality DNA extraction.
Understanding these sample-specific challenges and implementing appropriate extraction strategies is essential for achieving reliable results in molecular biology workflows. This guide examines the distinct issues associated with various sample types and offers practical solutions to overcome them, ensuring successful DNA isolation and subsequent analysis.
Blood and bodily fluids
Blood, saliva, urine, and other bodily fluid samples typically contain inhibitors like heme or mucin that can hinder downstream molecular applications like sequencing and PCR.
To extract DNA from these sample types, it’s important to consider workflow steps like heat and mixing that support a forceful but focused lysis and digestion – this will break open the cells without damaging DNA.
For applications like transplant diagnostics (e.g. identifying human leukocyte antigen [HLA] genes) or next-generation sequencing (NGS), sample analysis consistency and volume is key. Magnetic bead workflows can help increase sample throughput and uniformity to encourage high-quality genomic DNA results.
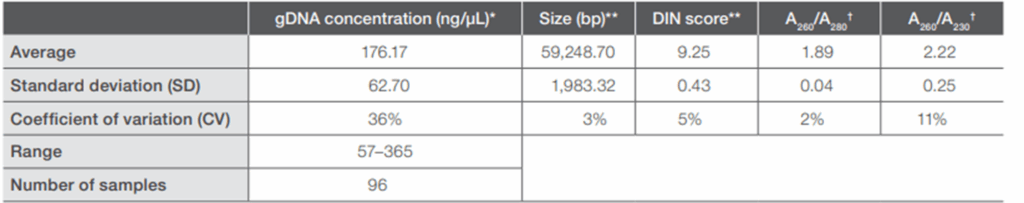
Tissue samples
Lysis efficiency depends on distinct, tissue-specific cellular structures in samples like liver, muscle, heart, brain, and other tissues. Tissues are often fibrous and tough, with rigid cell walls that necessitate mechanical disruption through homogenization or bead beating.
Homogenizers, like the Fisherbrand™ 850 Homogenizer, will typically yield higher DNA quantities than bead beating methods. If a hand-homogenizer is not available, researchers can opt for careful bead beating – rough supernatant mixing can cause a foamy homogenate that might be difficult to transfer into sample isolation. Incorporating RNase treatment can also help reduce RNA contamination within samples.
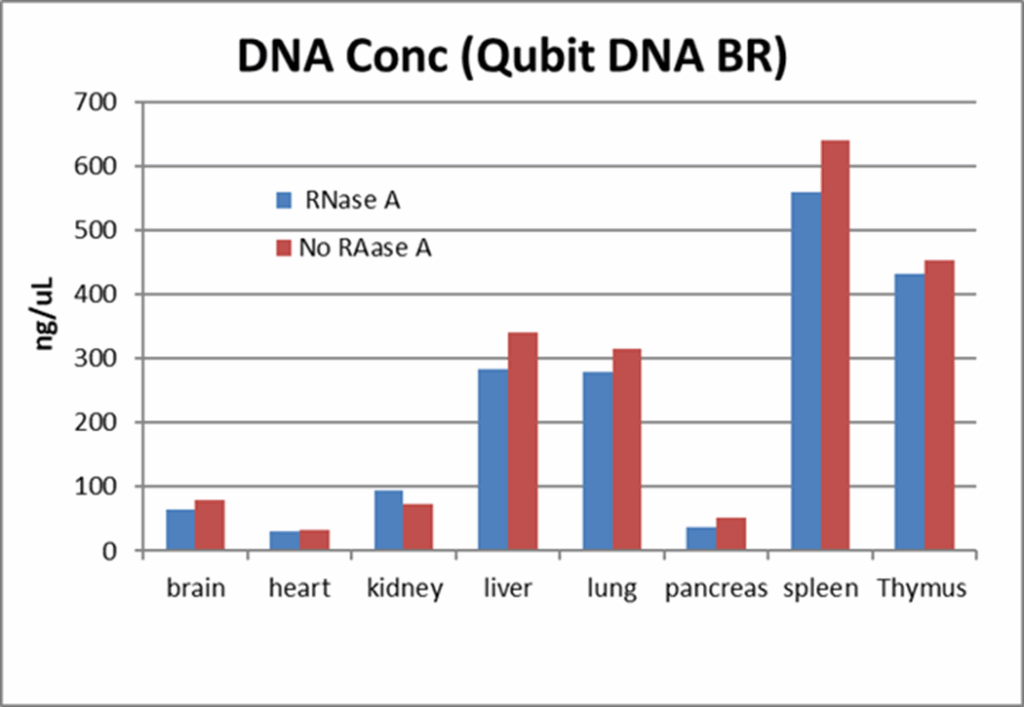
Incorporating RNase treatment can also help reduce RNA contamination within samples. The figure below illustrates comparative DNA yields from mouse brain, heart, kidney, liver, lung, pancreas, spleen, and thymus tissues isolated using MagMAX DNA Multi-Sample Ultra 2.0 chemistry with Applied Biosystems™ MagMAX™ Cell and Tissue DNA Extraction Buffer on a KingFisher instrument, both with and without RNase A treatment.
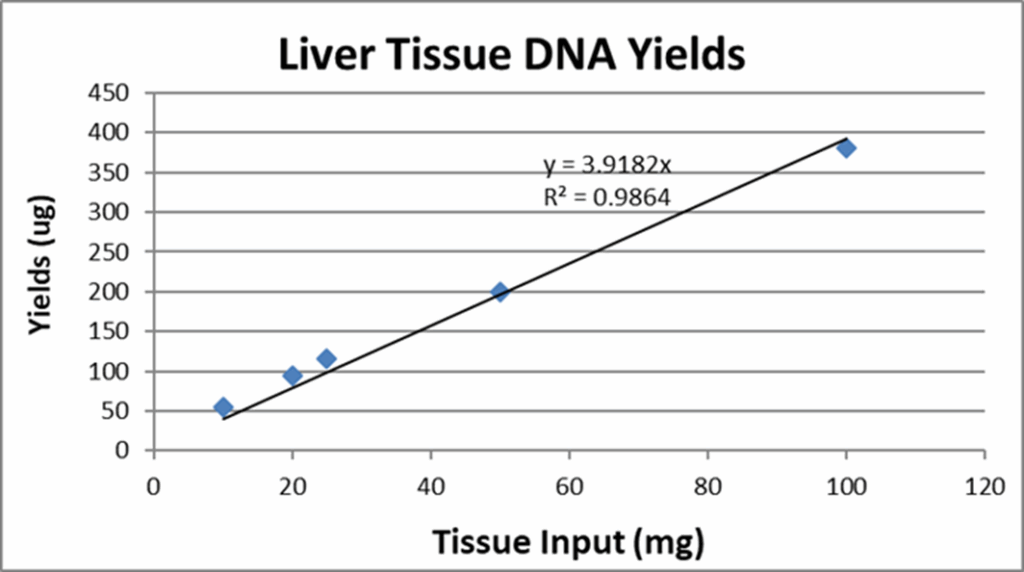
In general, the more tissue you put into an extraction, the more yield you will get. Figure 4 details mouse liver tissue isolated at varying sample inputs corresponding to their total yields. Typical quality absorbance ratios of A260/A280 tend to be greater than or equal to 1.6 on average.
Cell cultures
When isolating DNA from cell cultures, you will need to track cell viability, concentration, and maintain optimal density throughout expansion. For T cells specifically, you should carefully control culture conditions and perform regular quality checks.
Using specialized reagents like Gibco™ CTS™ OpTmizer™ T Cell Expansion SFM and Invitrogen™ Dynabeads™ Human T-Activator CD3/CD28 may help improve your process. Tools like the Countess II FL Automated Cell Counter help monitor your cultures effectively for reliable DNA isolation.
For more details on quality control during cell expansion, check out the application note “Cell Culture Quality Control During T Cell Expansion”.
Buccal swabs or dry swabs
Buccal swabs and dry swabs can be challenging because bacterial growth often occurs during storage. Oral microbiomes, skin, and other sample sources contain high concentrations of bacteria and contaminants, making proper drying, storage, and extraction protocols critical.
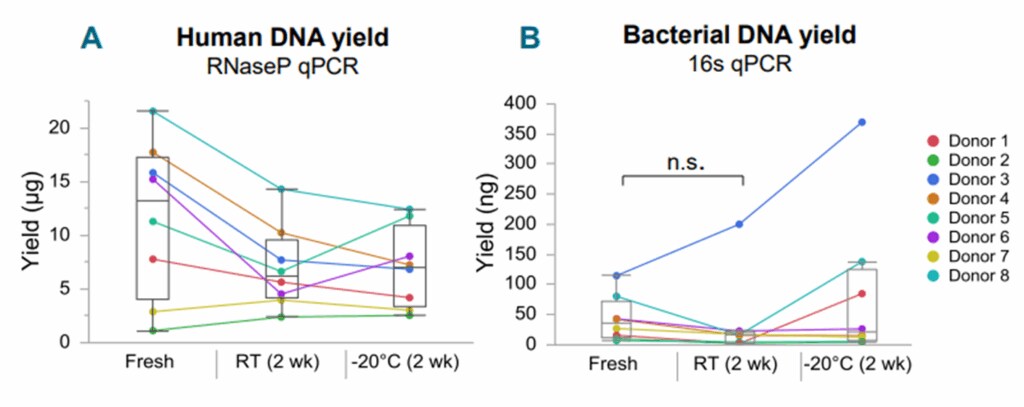
Magnetic bead-based methods with optimized chemistries can effectively target genomic and microbial DNA while removing sample-specific inhibitors (Figure 5). Even with excellent chemistry, you should plan to follow best practices for buccal swab isolations (Figure 6).
Consider using two swabs in a single isolation to double your yield. Extending the lysis stage can also improve recovery.

Samples in media (stool, swabs, saliva)
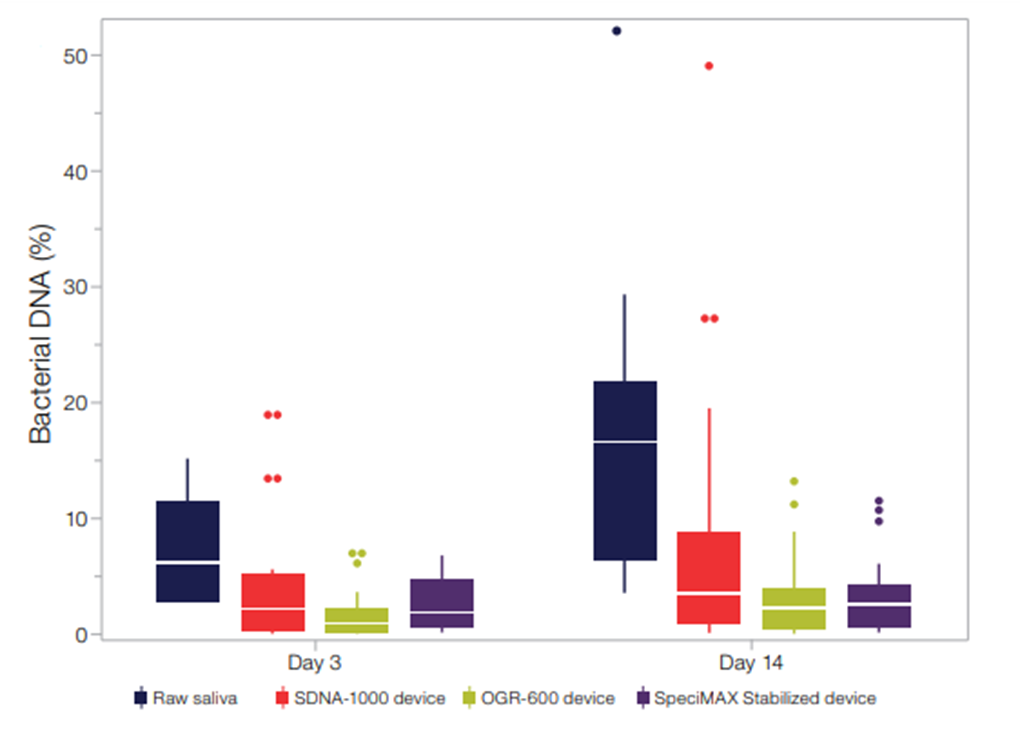
Stabilization media helps preserve sample integrity by preventing degradation from environmental factors, bacterial growth, and inhibitors. These media extend storage shelf life and improve experimental consistency. For saliva samples, stabilized collection devices effectively preserve genomic markers like APOE for GWAS studies. Research shows these devices reduce bacterial DNA content over storage time compared to raw saliva samples.
When you dilute samples like stool, saliva, and swabs in stabilization media, you will likely get lower DNA yields compared to smaller volumes of non-stabilized materials. Plan for flexibility in your sample inputs so you can adjust volumes to meet the DNA yield requirements of your downstream assays.
Plant samples
Isolating DNA from plants is challenging because of their tough cell walls and secondary metabolites like polysaccharides and polyphenols that can contaminate your DNA and interfere with downstream applications.
You can opt for specialized kits like the Thermo Scientific™ GeneJET™ Plant Genomic DNA Purification Kit (14) or Thermo Scientific™ MagMAX™ Plant DNA Isolation Kit (15) that incorporate PVP to reduce polyphenol inhibition. You can also modify plant workflows for automation by adapting PVP techniques to guanidinium-based solutions, as demonstrated in the webinar “High-Throughput automation in a tree disease research lab – A molecular technician’s journey.”
Formalin-fixed paraffin-embedded (FFPE) samples
FFPE samples are among the most challenging for DNA isolation because the formalin-fixing and paraffin-embedding require dewaxing and deparaffinization steps before you can access the nucleic acids. Traditional methods using multiple xylene and ethanol washes are environmentally harmful and tedious. Consider automated alternatives like the Applied Biosystems™ AutoLys M Tubes and Caps for deparaffinization, combined with Applied Biosystems™ MagMAX™ FFPE DNA/RNA Isolation chemistry. These methods use heating steps and proteinase digestion instead of harmful chemicals, significantly streamlining your workflow.

Streamlining different workflows into a single solution with automation
You may consider streamlining workflows across different sample types to save time and resources.
For manual work, consider TRI Reagent® when you need RNA, DNA, and proteins from a single sample. For automation, consolidate with bead-based kits like MagMAX DNA Multi-Sample Ultra 2.0, which can process whole blood, bone marrow, and saliva using a single KingFisher protocol. Sequential isolation kits like Applied Biosystems™ MagMAX Sequential DNA/RNA Kit let you extract both DNA and RNA from one sample. Also consider how sample preparation differs by type—raw stool samples require mechanical homogenization to release cells, while stool swabs in media might not need this step for microbial DNA identification. Similar streamlining can be applied to urine, whole blood, saliva, and other sample types.
Summary
With numerous DNA extraction options available, understanding the key variables will help you select the right method for your specific application. Consider the following factors to guide your decisions and support your research across workflows that require DNA extraction as an initial step.
- Sample volume input
- Sample concentration/yield requirements
- Sample type
- Sample throughput need
- Target analyte
- Downstream application
- Need for streamlining capabilities
More DNA extraction learning and resources
- Explore: Genomic DNA extraction and isolation
- Learn: DNA and RNA extraction and purification
- Learn: Spin columns in extraction
Protocols and application notes
- Protocol: Extraction of DNA using DNAzol reagent
- App note: Targeting gastrointestinal microbes from fecal and rectal swab specimens without bead beating
- App note: Analysis of concordance of genetic variants across sample types
- Tech note: RNA, DNA, and protein from a single sample
- App note: A convenient, solvent-free deparaffinization method for FFPE sample preparation
- Poster: Effects of buccal swab storage conditions on host and microbial nucleic acid stability and quality
- App note: Cell culture quality control during T cell expansion
- App note: Versatile solutions for human leukocyte antigen testing with peripheral blood
- App note: A comparative study of saliva collection devices using Axiom human genotyping arrays
- App note: Advancing hematological research with the MagMAX Sequential DNA/RNA kit
##
For Research Use Only. Not for use in diagnostic procedures.
© 2025 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.





Leave a Reply