
Gibson Assembly®, or Gibson cloning, is a powerful molecular cloning technique and popular alternative to the traditional restriction enzyme method.
Via a single tube isothermal reaction, the Gibson Assembly enables the seamless joining of DNA fragments. It is especially useful for building large or complex constructs from multiple segments, and it can be faster than other cloning methods.
Whether you’re a seasoned researcher or a newcomer to the Gibson Assembly protocol, this guide compiles learnings from our expert R&D scientists in molecular biology.
Table of contents
What is Gibson assembly?
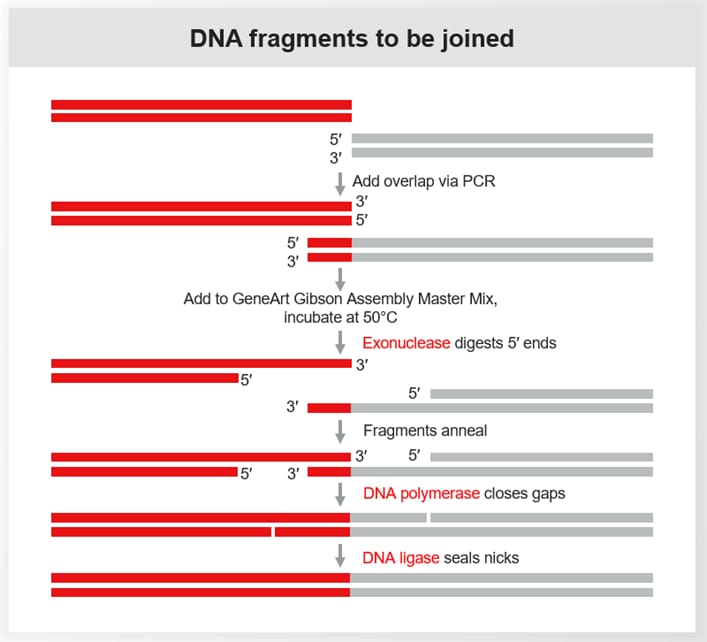
Gibson Assembly is a powerful method for DNA assembly that relies on the three enzymes exonuclease, DNA polymerase, and DNA ligase.
The process involves four main stages:
- Exonuclease Treatment: This enzyme chews back the 5′ ends of the DNA fragments, creating single-stranded overhangs.
- Annealing: Complementary regions of the DNA fragments anneal to each other.
- DNA Polymerase Extension: DNA polymerase fills in the gaps.
- Ligation: DNA ligase seals the nicks in the DNA backbone, resulting in a contiguous DNA molecule.
You might imagine each DNA fragment as a scrap of wallpaper or gift wrap; to join two scraps together without a visible seam, you could look for areas of exact pattern overlap at the cut edges and glue them together.
In this analogy, the single-strand overhangs at each DNA fragment’s end are like the pattern’s edges – as cut by exonuclease – and the annealing process is like the process of finding a visual overlap. Polymerase and ligation seal the fragment in place without any visible signs, or scarring.
When and why to use Gibson Assembly for molecular cloning
The Gibson Assembly technique has a few advantages over traditional restriction enzyme cloning, including:
- Flexibility to join multiple DNA fragments simultaneously, up to 6 fragments, regardless of their sequences, without specified restriction sites
- Precise and seamless union of fragments without scarring
- Takes fewer steps and less time than restriction enzyme cloning – usually under an hour
There are many practical applications for Gibson cloning. It can be used in seamless plasmid construction, synthetic gene and genome assembly, CRISPR vector creation, site-directed mutagenesis, modular DNA part assembly, and viral vector development for gene therapy and research.

Gibson Assembly protocol
The Gibson Assembly cloning method is a single-step isothermal process and often demands less than an hour of hands-on protocol time, depending on the number of DNA fragment inserts involved.
There are four key protocol steps.
1. Obtain DNA fragments
The success of Gibson Assembly depends heavily on the design of your DNA fragments and overlapping regions.
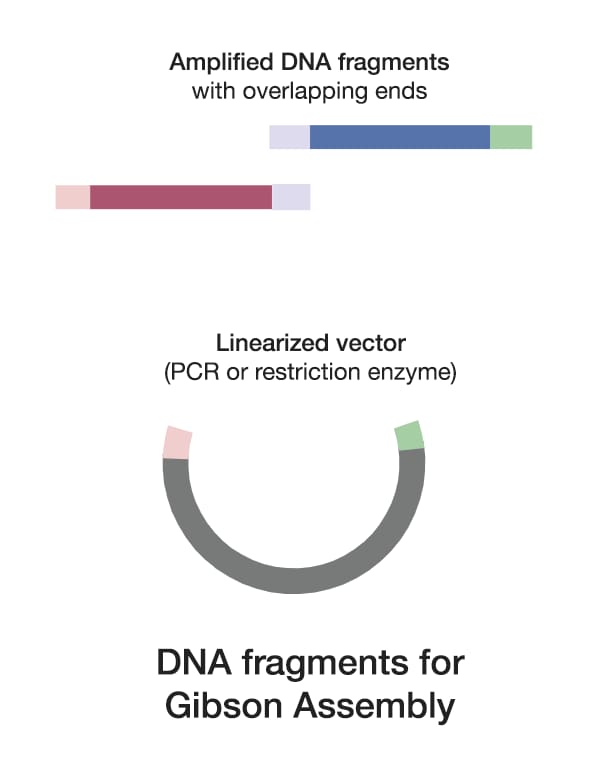
Design overlapping sequences carefully.
Aim for overlaps that are 20-40 base pairs long, ensuring they have a high GC content to promote stable annealing. Use software tools like SnapGene™ to design these overlaps accurately.
Optimize PCR conditions.
To ensure high-quality PCR products, optimize your PCR conditions for each DNA fragment. Use high-fidelity DNA polymerases, such as Platinum™ SuperFi II PCR Master Mixes, to minimize errors and ensure the production of clean, specific fragments. Purify your PCR products to obtain clean DNR fragments and verify the integrity of your PCR products via gel electrophoresis before proceeding
Linearize the vector of choice
Choose an appropriate vector for your assembly and linearize it using restriction enzymes or PCR. Linearization is crucial as it provides the necessary ends for the fragments to anneal and be ligated. When using a circular plasmid DNA as a template, use DpnI to reduce the template background after transformation.
2. Perform your Gibson reaction
To achieve successful Gibson Assembly, it is crucial to meticulously optimize the reaction conditions. Use a kit such as GeneArt™ Gibson Assembly™ HiFi Master Mix.
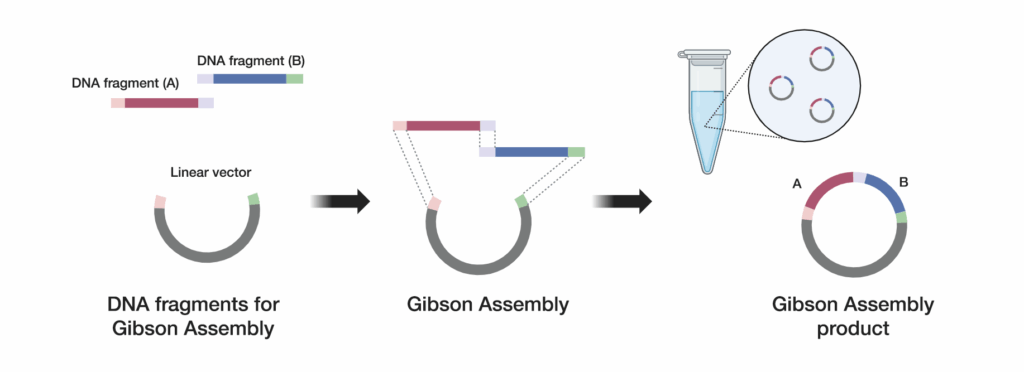
Combine your designed DNA fragments and linearized vector carefully
Ensure that the quantities of each component in a single-tube reaction are accurately calculated and properly dispensed. This attention to detail will help achieve high-efficiency assembly of your DNA fragments.
Include control reactions
Always include positive and negative controls in your experiments. A positive control confirms that the assembly reaction conditions are working, while a negative control (lacking one or more DNA fragments) helps identify potential contamination or non-specific assembly.
3. Transform competent cells
Use high-efficiency competent cells, such as One Shot™ TOP10 Chemically Competent E. coli, to maximize the number of successful transformants. Proper incubation times and temperatures are essential to achieve optimal results.
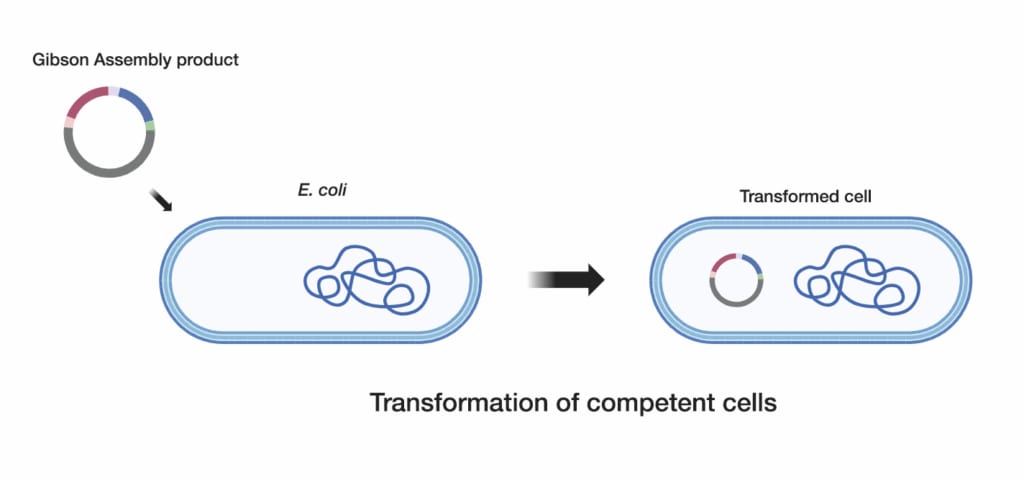
Plate the transformation mix on the appropriate plates.
Plate the transformed cells on selective LB agar plates to isolate individual colonies. This step is critical to ensure that only the cells containing the correctly assembled DNA construct will grow.
4. Screen colonies for construct
Screen several colonies to verify the correct assembly of your DNA construct. Use colony PCR, restriction digestion, or sequencing to confirm the presence and accuracy of the assembled DNA. Once verified, isolate and propagate the correct clone for further experiments.
Gibson Assembly troubleshooting and common questions
Below, our R&D experts share their responses to some of the most common Gibson cloning questions they hear from researchers.
1. How can I speed up the assembly process?
Researchers can speed up the assembly process by using unpurified PCR products, shortening reaction time, or using a rapid transformation protocol.
Using unpurified PCR products
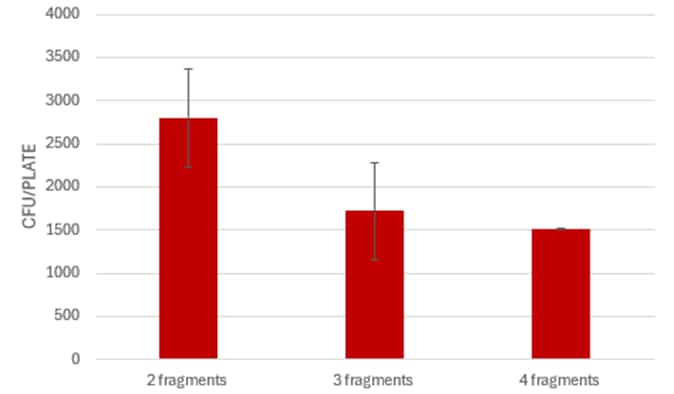
For a quicker assembly process, consider performing the Gibson Assembly directly from PCR products without purification. Simply collect up to 4 µL of each fragment and proceed with the assembly. This streamlined approach can save time while still yielding effective results.
Shortening reaction time
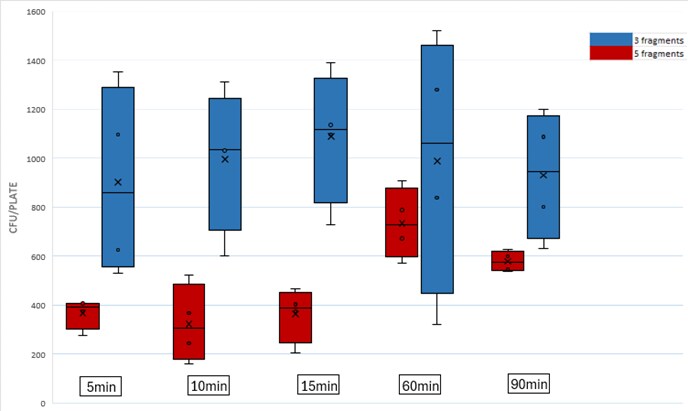
To save time, you can shorten the assembly duration. The protocol time is optimized for different assemblies, ensuring that the yield will not be significantly affected. This streamlined approach not only saves time but also maintains effective results, making your workflow more efficient.
Using a rapid protocol for transformation

For those looking to expedite the transformation process, consider using a rapid protocol. This method involves shortening the incubation times and using optimized reagents specifically designed for quick transformations. Simply mix the competent cells with the DNA and plate them directly onto selective LB agar plates without heat shock or recovery steps.
This approach is particularly effective for transformations using ampicillin-selective plates. The rapid protocol can significantly reduce the time needed while maintaining transformation efficiency with minimal changes.
2. Golden gate assembly vs Gibson Assembly: what’s the difference, and which should I use for cloning?
When it comes to DNA cloning, choosing the right assembly method can significantly impact your experimental success. Golden Gate Assembly and Gibson Assembly are two popular techniques, each with its own advantages.
Golden Gate Assembly utilizes type IIS restriction enzymes to create seamless DNA constructs, making it highly efficient for repetitive cloning tasks. On the other hand, Gibson Assembly employs a mix of enzymes to join multiple DNA fragments in a single, isothermal reaction, allowing for the assembly of larger and more complex constructs.
If your project involves modular cloning with high precision, Golden Gate might be the preferred choice. However, for assembling large DNA fragments or multiple inserts simultaneously, Gibson Assembly could be more suitable. Understanding the specific needs of your cloning project will help you decide which method to employ.
3. How do I design Gibson Assembly primers?
Designing primers for Gibson Assembly involves creating primers with overlapping sequences that match the ends of the DNA fragments you wish to join. Here are the key steps:
- Identify Overlap Regions: Determine the 20-40 bp overlap sequences needed for each fragment.
- Design Primer Sequences: Ensure that each primer contains the overlap sequence at the 5′ end and a region complementary to the target DNA at the 3′ end.
- Check Melting Temperatures: Verify that the melting temperatures (Tm) of your primers are compatible, ideally within 2-3°C of each other. Selecting overlapping regions of homology with Tm >50°C can improve efficiency.
- Avoid Secondary Structures: Use software tools to ensure your primers do not form hairpins or dimers. Following these steps will help you design effective primers for a successful Gibson Assembly reaction.
4. What is the proper Gibson Assembly overlap length for DNA fragments?
The proper overlap length for Gibson Assembly typically ranges from 20 to 40 base pairs. This length is sufficient to ensure that the fragments anneal specifically and stably to each other.
Overlaps shorter than 20 bp may not provide enough specificity and stability, leading to inefficient assembly, while overlaps longer than 40 bp may not significantly enhance the efficiency and can make the primer design more complex. Therefore, maintaining an overlap length within this range is crucial for the success of the Gibson Assembly process.
Gibson isothermal assembly: a convenient and fast alternative cloning option
Gibson Assembly is a versatile and efficient method for DNA assembly that can be mastered with careful planning and attention to detail. By following essential tips, you can optimize your protocols and achieve reliable, high-quality results in your cloning experiments.
Related Gibson Assembly resources and technologies
- Product: GeneArt Gibson Assembly HiFi Cloning Kit
- Protocol: Site-directed mutagenesis using the GeneArt Gibson Assembly HiFi Cloning Kit
- Whitepaper: From DNA contruction to protein in 1 day using a cell-free Gibson Assembly and rolling circle amplification workflow
- Learning Page: DNA Cloning Tips – Build clones with DNA fragments using Gibson Assembly
- Whitepaper: Utilizing both homology and nucleotide stitching techniques to build large constructs
##
© 2025 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. SnapGene is a trademark of GSL Biotech LLC. Gibson Assembly is a trademark of Telesis Bio Inc.
For Research Use Only. Not for use in diagnostic procedures.




Leave a Reply