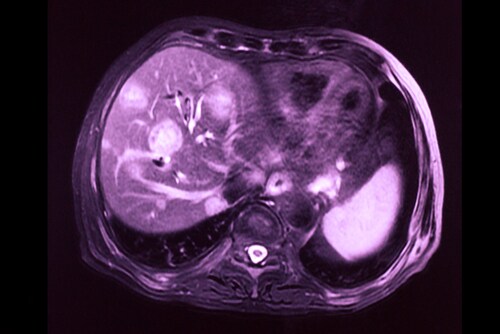
 Hepatocellular carcinoma is one of the most common forms of liver cancer and has a poor prognosis. Local ablation therapy with transarterial chemoembolization (TACE), a method whereby the blood supply to the tumor is restricted, is unfortunately associated with cholangiocellular differentiation in hepatocellular carcinoma. A potential explanation for this is that TACE provides selection pressure in favor of this particular cell population, which may be resistant to TACE.1 In the absence of biomarkers to differentiate between hepatocellular carcinoma and cholangiocarcinoma tumors or any real understanding of the pathogenesis of these types of tumors, treatment is a challenge and options remain few.
Hepatocellular carcinoma is one of the most common forms of liver cancer and has a poor prognosis. Local ablation therapy with transarterial chemoembolization (TACE), a method whereby the blood supply to the tumor is restricted, is unfortunately associated with cholangiocellular differentiation in hepatocellular carcinoma. A potential explanation for this is that TACE provides selection pressure in favor of this particular cell population, which may be resistant to TACE.1 In the absence of biomarkers to differentiate between hepatocellular carcinoma and cholangiocarcinoma tumors or any real understanding of the pathogenesis of these types of tumors, treatment is a challenge and options remain few.
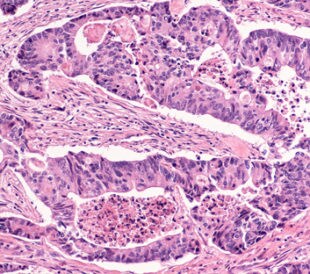
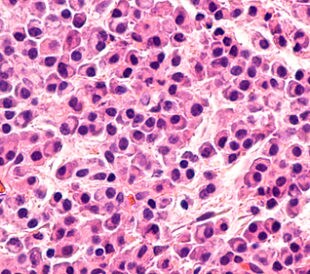
Building on their earlier research,1 Zen et al. recently profiled the protein expression of chemo-resistant, mixed-phenotype liver tumors to identify biomarkers for clinical use and better understand the pathogenesis of differentiation.2 The investigators used 55 human tissue samples from nine different types of liver tissue. The tissue types they used for differentiation were as follows: mixed hepatocellular carcinoma/cholangiocarcinoma after TACE (the researchers examined the areas of hepatocellular carcinoma and areas of cholangiocarcinoma separately), non-treated hepatocellular carcinoma, normal liver parenchyma, normal bile duct, non-treated peripheral cholangiocarcinoma, non-treated hilar/perihilar cholangiocarcinoma, PSC-associated hilar cholangiocarcinoma, and metastatic colorectal cancer.
The investigators prepared the tissue in 10-μm thick sections from formalin-fixed, paraffin-embedded tissue blocks before extracting the proteins using centrifugation at 14,000 × g at 4 ºC. Using 1D electrophoresis gels, they then separated out the proteins by molecular weight and excised the protein band that was visible at 4–20% (v/w) from each lane. Zen et al. took the products post-trypsin digestion and acquired mass spectra using liquid chromatography–tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap Velos hybrid mass spectrometer (Thermo Scientific).
In total, Zen and team identified 2,864 proteins, of which 2,628 (92%) had at least one unique peptide sequence and 2,009 (70%) of proteins had unique peptide sequences only. They used hierarchical clustering to build a class hierarchy for tissue types in relation to normalized protein values. The hepatocellular carcinoma and cholangiocarcinoma components of tumors post-TACE had different protein profiles. The research showed there were 95 proteins that were significantly modulated in the post-TACE hepatocellular carcinoma regions when compared with the cholangiocarcinoma regions. Seventy-eight proteins were up-regulated in hepatocellular carcinoma samples, as compared to 17 up-regulated proteins in cholangiocarcinoma tissues.
Although the study did not define the molecular mechanisms behind TACE-induced differentiation in hepatocellular carcinoma, it did identify protein sets specific to the hepatocellular carcinoma and cholangiocarcinoma phenotype of post-TACE mixed tumors. In performing this study, Zen et al. have identified potential biomarkers that can be used in the differential diagnosis of primary liver tumors.
References
1. Zen, C. et al. (2011) “Mixed phenotype hepatocellular carcinoma after transarterial chemoembolization and liver transplantation,” Liver Transplantation, 17(8) (pp. 943–954).
2. Zen, Y. et al. (2013) “Protein expression profiles of chemo-resistant mixed phenotype liver tumors using laser microdissection and LC–MS/MS proteomics,” EuPA Open Proteomics, 1 (pp. 38–47) .
Post Author: Miriam Pollak. Miriam specialised in neuroscience as an undergraduate but traded in lab work for a post graduate degree in science communication.
She has since had a career that has spanned science communication, science education and communications management.
However, Miriam has found her bliss balancing her love of writing and disseminating medical research with managing a multimillion dollar research budget for a childhood cancer charity in Australia.


Leave a Reply