Historically, cryo-electron microscopy (cryo-EM) structure determination required operators to collect one dataset after another. But with the onset of the SARS-CoV-2 pandemic and a subsequent queue of variants continuously impacting our ability to prevent infection with existing solutions, there’s a clear need for faster, more efficient, data collection for the purpose of drug discovery. In collaboration with Takeda Pharmaceuticals and Utrecht University, we demonstrate the power of high-throughput cryo-EM for epitope mapping, including the potential for improved antibody therapeutics.
Cryo-EM for rapid epitope mapping
An epitope is a part of a molecule against which an antibody attaches itself.
Therapeutic antibodies targeting the SARS-CoV-2 spike protein can play an important role in reducing the burden of the pandemic by protecting vulnerable populations against severe and potentially life-threatening disease.
Epitope mapping of large antibody panels allows researchers to identify vulnerable sites on the spike protein, predict and interpret the effect of new mutations, and speed decision-making in selecting antibody combinations that target non-overlapping epitopes. Moreover, structural information about epitopes can help identify liabilities in drug discovery pipelines in response to new emerging variants.
Therefore, in collaboration with Takeda Pharmaceuticals and Utrecht University, we set out to determine whether cryo-EM permits rapid epitope mapping for a large antibody panel.
Our study launched in the wake of the Omicron variant and subsequent strains, which escape most neutralizing antibodies, as well as the realization that we need additional tools for long-lasting prophylaxis against SARS-COV-2 infections.
Newfound speed, quality, and automation of cryo-EM data collection
Robust grid preparation and screening are critical for determining structures from every grid, and successful grid preparation is intimately linked to protein quality and concentration.
To evaluate the robustness of our spike protein sample, and to ultimately gauge the suitability of our 12 complexes for structural determination, we used the new automation capabilities of Thermo Scientific EPU Multigrid software on our Glacios Cryo-TEM.
The twelve screened grids were then subjected to unattended EPU Multigrid data collection using our Krios G4 Cryo-TEM equipped with an E-CFEG, Selectris X Energy Filter, and Falcon 4 Direct Electron Detector.
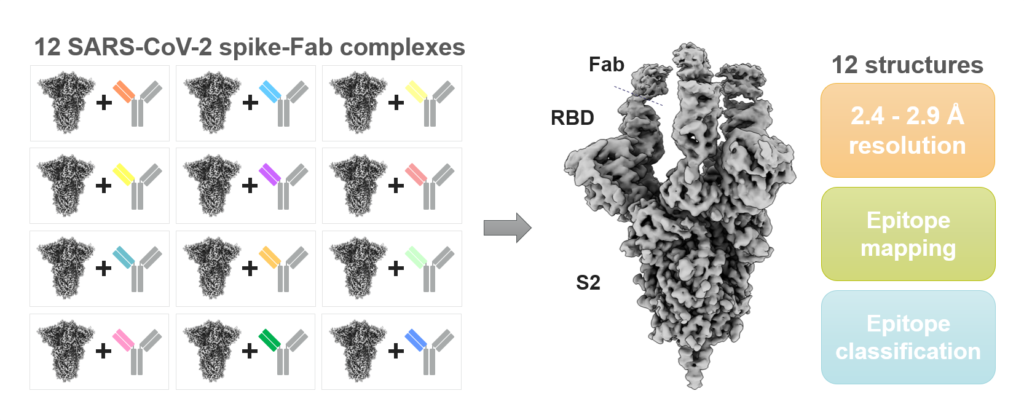
High-throughput cryo-EM epitope mapping of SARS-CoV-2 spike protein antibodies. Spike-Fab complex grids were imaged using Krios G4 cryo-TEM and EPU Multigrid, which resulted in twelve sub 3 Å cryo-EM reconstructions allowing epitope mapping and classification.
In a single 48 hour-long session, we set EPU Multigrid to collect 1,000 movies per grid, achieving a speed of 250 movies per hour.
This workflow enabled us to obtain 12 sub-3-angstrom structures of SARS-CoV-2 spike protein with different complexes and to determine their epitopes. Our results show the significant advances made in speed, quality, and automation of cryo-EM data collection.
“These exciting results show how state-of-the-art technology, and an optimized sample preparation workflow, can be used to accelerate antibody drug discovery, which is of great importance for COVID-19 and future pandemic threats,” said collaborator Daniel Hurdiss, Assistant Professor at Utrecht University.
Get an in-depth look at this workflow >>
We believe cryo-EM’s high-resolution revolution will soon be followed by a high-throughput revolution, which will improve existing applications through increased efficiency and reduced cost per dataset. It will also extend the range of cryo-EM applications to include fragment-based drug discovery and epitope mapping of large antibody panels.
//
Mazdak Radjainia is a Senior Staff Scientist at Thermo Fisher Scientific

Leave a Reply