DNA profiling has been used in forensics since 1985 when minisatellites and Southern blotting were the technology of the day.1,2 Sir Alec Jeffreys brought forth the idea of identifying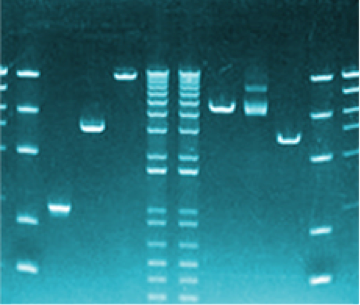 individuals based on their specific DNA characteristics and solved the first immigration case using this technique. Fast forward to the early 1990s when short tandem repeats (STRs) were introduced as a new polymorphic DNA marker type3,4 and have since become the gold standard for human identification in criminal casework, paternity and kinship analyses, and identification of missing persons5,6. From the original nine markers in the Applied Biosystems AmpFLSTR Blue, Green and Yellow PCR Amplification Kits to the newly instituted 20 core CODIS loci7 in the Applied Biosystems GlobalFiler PCR Amplification Kit, profiling capability has increased in its power of discrimination (PoD) and expanded international data sharing efforts.
individuals based on their specific DNA characteristics and solved the first immigration case using this technique. Fast forward to the early 1990s when short tandem repeats (STRs) were introduced as a new polymorphic DNA marker type3,4 and have since become the gold standard for human identification in criminal casework, paternity and kinship analyses, and identification of missing persons5,6. From the original nine markers in the Applied Biosystems AmpFLSTR Blue, Green and Yellow PCR Amplification Kits to the newly instituted 20 core CODIS loci7 in the Applied Biosystems GlobalFiler PCR Amplification Kit, profiling capability has increased in its power of discrimination (PoD) and expanded international data sharing efforts.
Currently allele classifications are identified by PCR-based fragment sizing by capillary electrophoresis (CE)5 following a simple nomenclature convention8-10. The alleles are called relative to known sized allelic ladders, with a number value indicative of the number of complete repeat motifs and any additional nucleotides8). This convention is based on the size variation produced by CE systems; but it does not account for any sequence-based differences between alleles of the same size that are the result of any number of genetic rearrangements. The PoD of these size-based alleles has proven to be sufficient to provide information on exclusion or inclusion for human identification purposes, and has grown with the introduction of large multiplexes7,11 such as the GlobalFiler PCR Amplification Kit and the NGM Detect Amplification Kit.
Next-generation sequencing (NGS) adds a new twist to the field of forensic human identification, providing benefits over CE analysis in terms of actual sequence context of the repeat motifs, the ability to multiplex even more markers, and the analysis of degraded samples12–14 which may not provide full profiles from CE analysis, especially for the larger loci. NGS has shown that STRscan be typed effectively and will produce profiles that are compatible with those of CE analyses, even from challenging crime scene samples15. The resulting STR profiles from NGS provide more information than just the allele number by analyzing the nucleotide sequence of the repeat motifs as well as nearby variations in the flanking regions. Evaluations of NGS-based STR typing show that it may be useful in routine casework as it may improve mixture resolution and increase the PoD12,16 by including both more loci, which are contained in the Precision ID GlobalFiler NGS STR Panel v2, and the additional sequence information.
While NGS opens to the door to greater discovery, one of the big questions is how much sequence variation exists in these STR loci. A recent study by Novroski et al of over 700 individuals from four population groups (African American, Caucasian, Hispanic, and Chinese) found that these three STRs in particular, D21S11, D2S1338 and D12S391, showed higher allelic diversity from sequencing the repeat motifs in all four populations. Several loci, CSF1PO, D10S1248, TH01 and TPOX, showed little to no diversity in length vs sequence based alleles.17 To figure out how to really use this data, more studies are in process and a new repository, POPSEQ, for collating these discoveries is being managed by North Carolina State University.
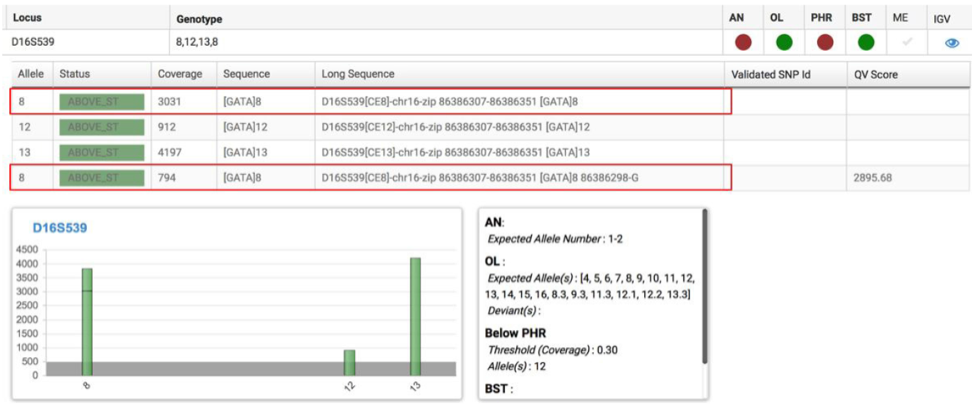
In addition, Single Nucleotide Polymorphisms (SNPs) in the flanking regions of these STR repeat motif are being investigated to determine how they can be used in resolving complex mixture and kinship analyses. D5S818, D7S820, D13S317 have multiple polymorphisms within haplotypes, and this has resulted in new alleles, based on variation in these flanking regions and they are associated with multiple populations and allele sizes. D16S539 shows a single SNP which is also well represented in the most common allele sizes and populations, making it useful for new allele determinations.18
STR analysis by NGS does pose new challenges to the DNA analyst. These new findings will affect how the data are analyzed, stored, and reported to law enforcement and the court system. And the forensic community will need to decide how and when this new information is added to national DNA databases. The current nomenclature is not able to take into account the additional variation. The executive board of the ISFG introduced a DNA commission to assess the considerations needed for sequence-based STR nomenclature. It will be critical that the new naming convention provides compatibility with the greater than 70 million profiles from CE-based STR typing that are housed in these national DNA databases. Presently, the forensic community expects that both CE- and NGS -based STR typing methods will continue to coexist.19
 CE-based STR analysis is able to resolve an estimated > 60% of casework samples. For the small percentage of cases that currently cannot be solved by CE the application of NGS-based STR analysis will depend on a number of lab, case and sample aspects, such as ease of use, the value of the increased PoD, complexity of the case and integrity of the samples. The continued evolution of NGS in forensic genetics may help make the world a safer place, provide justice to victims, and free the innocent.
CE-based STR analysis is able to resolve an estimated > 60% of casework samples. For the small percentage of cases that currently cannot be solved by CE the application of NGS-based STR analysis will depend on a number of lab, case and sample aspects, such as ease of use, the value of the increased PoD, complexity of the case and integrity of the samples. The continued evolution of NGS in forensic genetics may help make the world a safer place, provide justice to victims, and free the innocent.
For Research, Forensic, or Paternity Use Only. Not for use in diagnostic procedures.
References:
- P. Gill, A. J. Jeffreys, D. J. Werrett, Forensic application of DNA ‘fingerprints’, Nature 318 (6046) (1985) 577-9.
- A. J. Jeffreys, V. Wilson, S. L. Thein, Individual-specific ‘fingerprints’ of human DNA, Nature 316 (6023) (1985) 76-9.
- C. Puers, H.A. Hammond, L. Jin, C.T. Caskey, J.W. Schumm, Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01[AATG]n and reassignment of alleles in population analysis by using a locus-specific allelic ladder, Am. J. Hum. Genet. 53 (1993) 953–958.
- P. Gill, C. Kimpton, E. D’Aloja, J.F. Andersen, W. Bar, B. Brinkmann, et al., Report of the European DNA profiling group (EDNAP)—towards standardisation of short tandem repeat (STR) loci, Forensic Sci. Int. 65 (1994) 51–59.
- P. Gill, R. Sparkes, C. Kimpton, Development of guidelines to designate alleles using an STR multiplex system, Forensic Sci. Int. 89 (1997) 185–197.
- B. Budowle, T.R. Moretti, A.L. Baumstark, D.A. Defenbaugh, K.M. Keys, Population data on the thirteen CODIS core short tandem repeat loci in African Americans, U.S., Caucasians, Hispanics, Bahamians, Jamaicans, and Trinidadians, J. Forensic Sci. 44 (1999) 1277–1286.
- D. R. Hares, Selection and implementation of expanded CODIS core loci in the United States, Forensic Sci Int Genet 17 (2015) 33-4.
- W. Bär, B. Brinkmann, B. Budowle, A. Carracedo, P. Gill, P. Lincoln, et al., DNA recommendations. Further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics, Int. J. Legal Med. 110 (1997) 175–176.
- J.M. Butler, Genetics and genomics of core short tandem repeat loci used in human identity testing, J. Forensic Sci. 51 (2006) 253–265.
- P.M. Schneider, Scientific standards for studies in forensic genetics, Forensic Sci. Int. 165 (2007) 238–243.
- P.M. Schneider, Expansion of the European Standard Set of DNA database loci— the current situation, Profiles DNA (2009) 6–7.
- C. Borsting, N. Morling, Next generation sequencing and its applications in forensic genetics, Forensic Sci. Int. Genet. 18 (2015) 78–89.
- W. Parson, G. Huber, L. Moreno, M.B. Madel, M.D. Brandhagen, S. Nagl, et al., Massively parallel sequencing of complete mitochondrial genomes from hair shaft samples, Forensic Sci. Int. Genet. 15 (2015) 8–15.
- M. Eduardoff, C. Santos, M. de la Puente, T.E. Gross, M. Fondevila, C. Strobl, et al., Inter-laboratory evaluation of SNP-based forensic identification by massively parallel sequencing using the Ion PGM, Forensic Sci. Int. Genet. 17 (2015) 110– 121.
- M. Scheible, O. Loreille, R. Just, J. Irwin, Short tandem repeat typing on the 454 platform: strategies and considerations for targeted sequencing of common forensic markers, Forensic Sci. Int. Genet. 12 (2014) 107–119.
- K.B. Gettings, R.A. Aponte, P.M. Vallone, J.M. Butler, STR allele sequence variation: current knowledge and future issues, Forensic Sci. Int. Genet. 18 (2015) 118–130.
- N.M.M. Novroski et al. Characterization of genetic sequence variation of 58 STR loci in four major population groups Forensic Science International: Genetics 25 (2016) 214–226
- K.B. Gettings et al. The next dimension in STR sequencing: Polymorphisms in flanking regions and their allelic associations Forensic Science International: Genetics Supplement Series 5 (2015) e121–e123
- W. Parson et al. Massively parallel sequencing of forensic STRs: Considerations of the DNA commission of the International Society for Forensic Genetics (ISFG) on minimal nomenclature requirements Forensic Science International: Genetics 22 (2016) 54–63




Leave a Reply