Dr. Tim Inglis shares with us his findings on FAST methods of antibiotic resistance detection and most recently, on flow cytometry-assisted antimicrobial susceptibility testing using the Attune NxT Flow Cytometer.
 Tim Inglis, Ph.D.
Tim Inglis, Ph.D.
Dr. Tim Inglis works at the School of Medicine at the University of Western Australia and the School of Biomedical Sciences where his research team is based. He also works at the state pathology service, PathWest Laboratory Medicine, all located on the Queen Elizabeth II Medical Campus. Dr. Inglis’ research is principally concerned with sepsis and antimicrobial resistance.
What drew you to this area of research?
What drew me to this area of research goes back to the mid 1980s when a colleague of mine was working with a very early flow cytometer on the aggregation of Staphylococci in liquids. Then since that, I’ve had a longstanding interest in applications of flow cytometry to bacteriology.
You have recently published on a new method. Can you describe your publication?
The new method group we published on is known as FAST. It’s flow cytometry-assisted antimicrobial susceptibility testing or FAST for short. It uses an acoustic flow cytometer to rapidly determine the response of bacteria to antibiotics they’re exposed to.
Can you describe the significance of this?
The significance is a combination of two things. It’s principally about achieving the same accuracy as the standard methods that are used in clinical laboratories around the world, but doing so in a much shorter time because severe and invasive bacterial infections are time-critical medical emergencies. No doctor is going to want to wait two to three days for an answer that he or she could get in a matter of a few hours or even a few minutes. That’s why our FAST method is very important because it achieves accuracy in a much shorter period of time.
As far as the FAST methodology, what are your next steps?
There are three key components to this. One is to push for high throughput and automate so that handling of samples can be done on a much larger scale as a clinical laboratory would expect. We obviously also need to expand our repertoire of drug/bug combinations, and that’s something we’re already working on. The third thing is to simplify the procedures and to get them into a form that can easily be used by technical staffing clinical laboratories.
Let’s talk about your flow cytometry workflow and how you use flow cytometry in your research.
Our initial use of flow cytometry in our research has been mainly focused on a discovery process. We’ve used the Attune NxT Flow Cytometer for us to discover how better to do susceptibility testing. That led us to specific discoveries about how we should prepare our specimens and bacteria, how we clean them up, how we expose them to the fluorescent dyes that are detected on different channels of the flow cytometer. Then the addition of the reagents, in this case, the antibiotics that give us the changes that can be detected by flow cytometry in a short period of time.
After the fluid handling process has been completed by the acoustic flow cytometer, we then obviously proceed to a data analysis step. That gives us the critical differentiation between susceptible and non-susceptible bacteria. We’ve also used a digital microscope to give us a useful extra set of subjective insights into the processes that are going on. This, for microbiologists, is very much sticking within our comfort zone because flow cytometry is not the traditional province of clinical microbiologists. Microscopes obviously are. Incorporating that in the discovery workflow has been an important part of where this has gone so far and is reflected in the first publication that my colleagues have produced on the FAST method.
In adopting flow cytometry, it’s not a traditional tool. Could you comment on how difficult or easy it was for you to start using flow cytometry and why you chose that method?
The difficulties of moving into flow cytometry have been something that I personally experienced since the mid 1980s. The initial equipment that was available then clearly wasn’t suitable for the task of susceptibility testing. It was only in the 1990s that a small number of experts started to propose that flow cytometry should be used for antimicrobial susceptibility testing. Even then, the accuracy of conventional hydrodynamic flow cytometers was not sufficient to produce reliable results in the kind of time frame that was going to make a difference clinically.
We’ve had to overcome some of those road bumps. The speed bumps that got in the way were accuracy in bacterial cell size range, also issues of background noise, and contamination with particulate material. Some of these obstacles to progress are familiar to those people who set up molecular diagnostics. They’re not the same, but they were a similar quality and magnitude of problems.
Since then, now that we’ve got those problems out of the way, we’re progressing much more quickly to expanding the test repertoire of bug/drug combinations that I mentioned earlier.
Can you comment on your use of the Attune NxT Flow Cytometer in your research and in the FAST method?
Our use of the Attune NxT Flow Cytometer, it’d be fair to say, is very much at front and center of where we’ve taken this development process. We wouldn’t have got to this area without acoustic flow cytometry. The additional accuracy of the Attune [NxT] has been a critical step in overcoming those principal obstacles to analyzing bacteria at a single cell level.
The [Attune] NxT has given us additional channels for much more specific analysis of different color dyes. Whilst we started with one-color dye with SYTO 9 and got some very good results with that, the options that were given as a result of being able to multiplex with more than one dye and on additional channels open up a much wider range of potential tests to it.
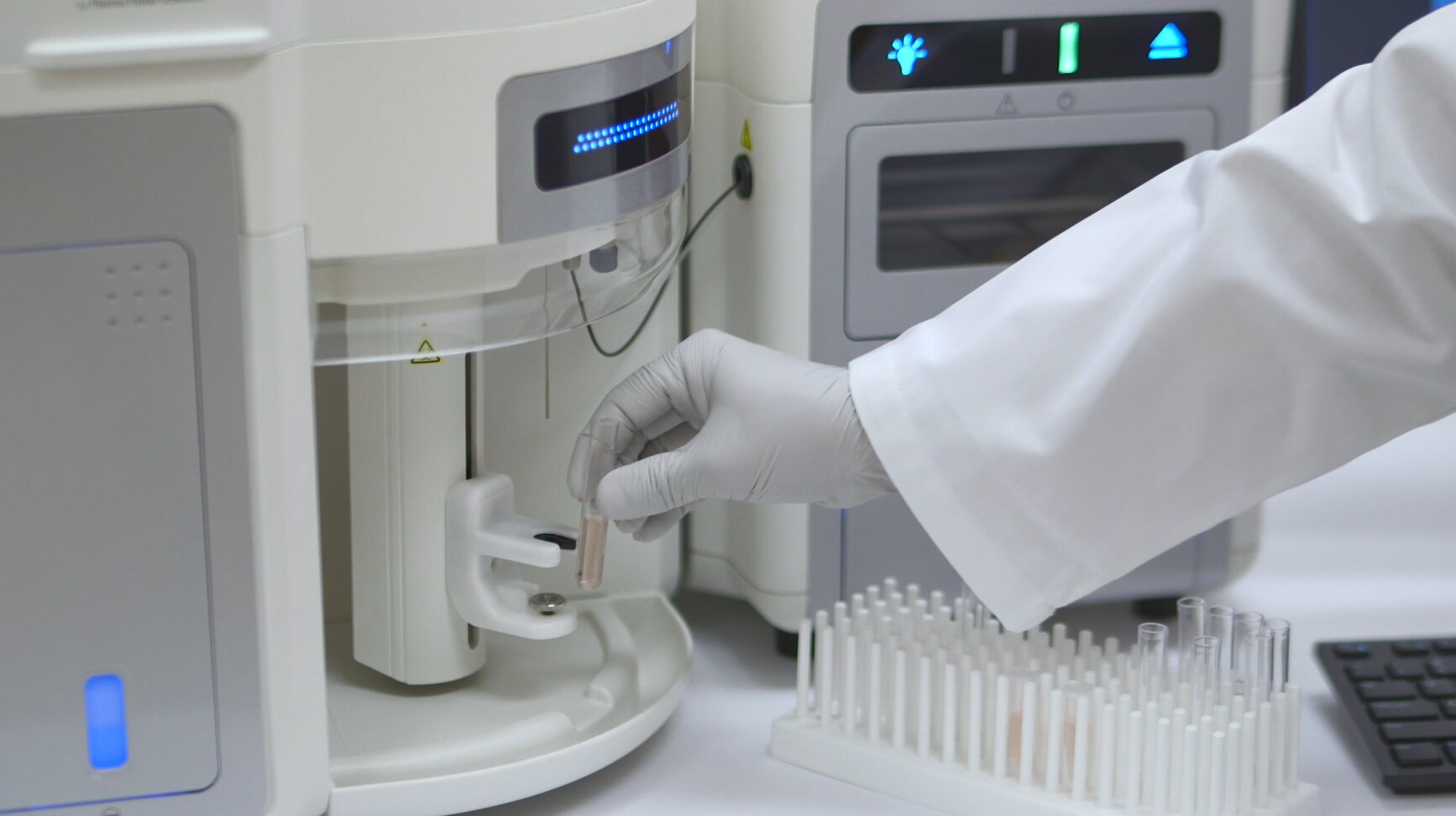
Would you consider the Attune NxT Flow Cytometer to be a significant breakthrough or innovation for you in your research?
The Attune NxT is clearly a significant breakthrough, and I can speak from something like three decades of on-and-off use. You could say even a love-hate affair with flow cytometry, a desire to use flow cytometry for clinical bacteriology but frustrated by the limitations of the technology.
The Attune NxT has given us an opportunity to analyze bacteria at a single cell level. And in combination with the range of vital dyes available to us, it’s expanded the potential repertoire of culture-independent bacteriology well beyond the boundaries that are set by molecular biology and in particular, by real-time PCR.
It really is beginning to look like bacterial cytomics are here to stay, and the Attune NxT has been the critical piece of technology.
Could you comment about your satisfaction with working with Thermo Fisher Scientific?
We’ve been very comfortable working with Thermo Fisher Scientific for the last four years. We have a collaboration arrangement that goes back over three and a half years and has taken us from initial, in some cases, accidental/ serendipitous discoveries, to a point where the technology we’re working on is being adopted by internationally respected reference laboratories overseas. That’s very unusual for a group based in somewhere as geographically isolated as Western Australia. This collaboration is a very unusual one, at least, for us locally and has been a very productive one.

Could you describe your workflow for the FAST method?
In essence, what we do is recover bacteria, suspend them in a suspension that can be analyzed with a minimum of background noise by the Attune [NxT] Flow Cytometer, and then we expose the bacteria to antibiotics. We treat the bacteria with a vital dye that is taken up and gives the flow cytometer the opportunity to see those bacteria.
We analyze using the software that’s preinstalled and generate a series of gated populations, and then we do further downstream analysis on those gated populations to understand the shift in the size, shape and color intensity of those bacterial populations as a response to antibiotics.
What we’re demonstrating with the Attune [NxT] Flow Cytometer is a change that is precipitated by exposure to different concentrations of antibiotics. This is very similar to but much faster than the conventional culture-based methods of MIC determination, minimum inhibitory concentration.
We believe that it’s much more accurate though because the end point is far more clearly determined and at the very least, is as accurate and probably, in some sense, is more accurate than conventional broth microdilution techniques.
What is the time difference of doing your FAST method as compared to a traditional method?
The time differential between FAST MIC and broth microdilution MIC, the international reference standard is approximately 18-24 hours at the moment. What would normally take for the international standard 24 hours takes us just less than three hours with manual fluid handling. Now some of that, of course, is as a result of exposure to antibiotic, time being incorporated into the process. The qualitative result that can be obtained, which is more akin to a break point susceptibility can be achieved in as little as less than 30 minutes. With automation, obviously, some of that manual fluid handling could be removed from the process. We anticipate at some point in the future that a leaner process will emerge out of this as standard operating procedure, if you like, which will take some of that time frame out of the overall workflow.
At the moment, we realize that we are best-in-class. We’re keen to remain ahead of the rest of the field and we do believe that there are some points in which we can further improve the system that we’re currently using.
Can you describe the type of samples that you’re testing?
The samples that we’re currently getting and testing are suspensions of bacteria, which, for reasons of meeting regulatory standards, have been recovered from stock culture collections. There is a standard ATCC set of criteria for recovering those organisms. One of my colleagues, Kieran Mulroney, has already presented at international meetings on application to a sterile fluid specimen from a subject with CAPD-peritonitis. We believe that there are a number of clinical applications for this where we could, at some point in the future, be looking at direct application of the FAST method to clinical specimens. For the moment, at least, in order to meet the validation standards and expectations, we’re working on primary isolates: initial isolates recovered on solid or in liquid media, washing those organisms, and then taking them through to a FAST susceptibility result.
It sounds like you’re very heavily involved in taking the basic research and translating that into a clinical assay. Could you discuss that briefly?
Certainly. Taking a discovery through to implementation and adoption in practice is often a very lengthy process. Taking an antimicrobial susceptibility test process from its baseline discovery is a new innovation to readiness in the clinical laboratories, restricted at certain stages by the expectations of regulatory agencies like the FDA and here, there’s an organization called the Therapeutic Goods Administration. We need to meet those regulatory standards, and that requires not only training the equipment to do the job that we need to do and convincing ourselves that this is the way to go, but an assay method has to be properly written up. It has to be validated with an extensive collection of representative bacterial isolates. That we’ve done for one drug/bug combination and have published on that. We’re in the process of working with colleagues who collaborated with us on that first publication to complete independent verification of the validation process that we have already undertaken. That then needs to be done for all of the drug/bug combinations that we work on.
You’ve got to have a real focus on the clinical benefits and the needs of doing this sort of work in a clinical laboratory environment, in a pathology laboratory service to understand why it’s so important to meet those validation standards.
It is a big task and it requires a very keen focus on that finish line that we are working towards, but as a clinical translational research group, that is very much part and parcel of what we do on this site.
What do you like most about your Attune NxT Flow Cytometer?
What I like most about the Attune NxT [Flow Cytometer] is its accuracy and its speed of processing. I could list a range of other things that I like about it because we are building up that list as we discover more aspects of its functions, but the core things are speed and accuracy that are not available with hydrodynamic flow cytometers.
Can you describe the advantages of the Attune NxT Flow Cytometer in your assays?
I guess the principal advantage of the Attune NxT [Flow Cytometer] has been accuracy in the bacterial particle size range, so the ability to do accurate cellular analysis across a range of different acquisition wave lengths. The ability therefore to generate the baseline data rapidly in a format that can easily be analyzed with third-party software to take things on to a much higher plane. In particular, for our FAST assays, the ability to quickly describe the unexposed, morphotype the UCM that was described in Mulroney and Hall, et al., the first paper on this technique.
What other clinical specimens do you work with?
We are working with a range of additional clinical specimens, and I think the most notable one that we are most excited about is blood cultures. Invasive infections require blood culture processing. A subject with sepsis will have a blood culture taken. Traditionally, this will take, in our center, a median of 18 hours before it flags positive. After collection it may take anywhere between two to three days before the susceptibility results become known.
We’ve run a pilot study using our FAST technique, and in that pilot study of 30 consecutive blood culture positives, all of the gram-negatives that we analyzed were correctly called by FAST.
Most importantly, this saves us at least 24 hours but more like two days; 48 hours before we get the definitive susceptibility result. We’ve been able to do this across a range of different intravenous antibiotics beyond the carbapenems that were first reported in our initial report on the FAST technique. We’re now confident that we can start applying this to clinical applications and particularly, to invasive infections.
If you bear in mind that every hour’s delay to starting the right antibiotic for a subject with bloodstream infection results in an 8% increase in mortality, you realize that that kind of advance is going to save lives.
Do you use the Autosampler in your workflow?
In the use of the [Attune] NxT, so far, most of our work has been done with manual fluid processing, but we have recently gained access to an Autosampler and we have begun to explore higher throughput processing. I anticipate that the use of auto sampling will enable clinical laboratories to process larger numbers of clinical isolates using a FAST or modified FAST method in the near future.
What are your hopes for this field of research in the future?
We could spend a long time going over my hopes for bacterial flow cytometry, but I’ve gone down on the record as, I guess, saying two key things. One is that we see the introduction of FAST or flow-assisted susceptibility testing; in its various different forms into clinical practice as quickly as possible because it is going to make a difference. It is going to cause a significant shift to the left on the susceptibility testing timeline. We know that that is critical in combating the disastrous rise of antimicrobial resistance (AMR). AMR is clearly a major global concern.
My second event horizon is the wider application of bacterial flow cytometry to culture-independent diagnostic tests. This is more than just antimicrobial susceptibility testing. This is going to be the application of flow-based methods to detecting the presence of bacteria, accurately enumerating those bacteria and identifying them as well as determining their antimicrobial susceptibilities. That is a dream that some of us started to chase rather boldly in the 1980s but without the tools to do so. We’re starting to see those tools come within our grasp. I look forward to the next five years, which could well see the development of bacterial cytomics, or bacterial flow cytometry, to a similar level as where molecular biology has gone in the field of infectious diseases diagnostics. Thank you.
This work is being performed in the FAST Lab; a collaboration between the UWA School of Biomedical Sciences, and PathWest Laboratory Medicine, supported by Lab Without Walls, Rotary, the Health Department of Western Australia, the Bill & Melinda Gates Foundation and NESTA, UK.
For Research Use Only. Not for use in diagnostic procedures.


I am a retired former asst. professor of Family Medicine in Louisville Ky. USA. I have been found to have a resident infection with MAP. Have been interested in flow cytometry follow up on my infection after 12 months on treatment. Your methods seem to offer a superior technique for bacterial analysis. Currently Dr. John Aitken in New Zealand has my blood culture isolate which showed a strong growth of MAP after about 4 weeks. Clinically I feel much better but have questions about adequacy of therapy. Do you have thoughts about this problem. Flow cytometry testing for bacterial infection is not very advanced here.
We tracking for salmonella cells extra and intracellular in small intestine. Our result confirme the flow cytometer gave microbial load with viability. The chance of AST analysis by flow cytometer is possible but if invasion new kits contains antibiotics labelled by fluorescence dyes to diagnose antibiotic vs. Bacteria.