“A comprehensive panel is essential to obtain a good yield and may be more efficient than whole exome sequencing,” says Alessandra Murgia, Director of Laboratory of Molecular Genetics and Associate Professor of Pediatrics at Department of Women’s and Children’s Health at the University of Padua, Italy.
“Gene coverage uniformity is important, but if we were to use a poor gene collection, the best coverage uniformity would not lead to the same result. What can make the difference is a very large repository that includes the best possible collection of pathogenic genes processed with high coverage uniformity—like that available in Ion AmpliSeq On-Demand.”
The advancement of targeted next-generation sequencing (NGS) has been instrumental in accelerating the pace of discovery for Murgia’s work in neurodevelopmental disorders (NDDs) research and testing.
Upcoming talk: “New approaches to human genetic diseases”
Register today for Oct. 10 talk, live (Amsterdam) or online
Dr. Murgia will discuss her pragmatic approach to the hunt for the genetic mutations that cause neurodevelopmental disorders and hereditary deafness: look in the most obvious places first. Her work focuses on understanding NDDs, intellectual disability/autism spectrum disorders, early onset epilepsy, and hereditary deafness.
NDDs are clinically and etiologically highly heterogeneous, and are characterized by a wide range of overlapping clinical features. These characteristics pose significant challenges for diagnoses based on exclusive clinical criteria.
The etiological background of these disorders is essential in gaining fundamental insights into the pathogenic mechanisms and pave the way to a fine understanding of different phenotypes and variable clinical expression associated with NDDs.
However, designing a custom panel for disease research requires hours of literature searches to select the genes most appropriate for their research, often without any information about how the panel will perform in their lab.
Murgia and her team were able to design custom panels of relevant, wet lab-verified genes by browsing disease research areas in the Ion AmpliSeq On-Demand catalog.
“For my areas of interest (neurodevelopmental disorders, hearing loss),” Murgia says, “we want to develop a comprehensive panel, not just an array of genes. The Ion AmpliSeq On-Demand gene repository and disease database leverages public information and computational analysis of publications.”
This enables us to quickly assemble relevant panels for the nervous system from intellectual disability to white matter disorders and epilepsy, and it gives us a head start in the upfront work related to gene panel building; namely the selection of genes to include,” she says.
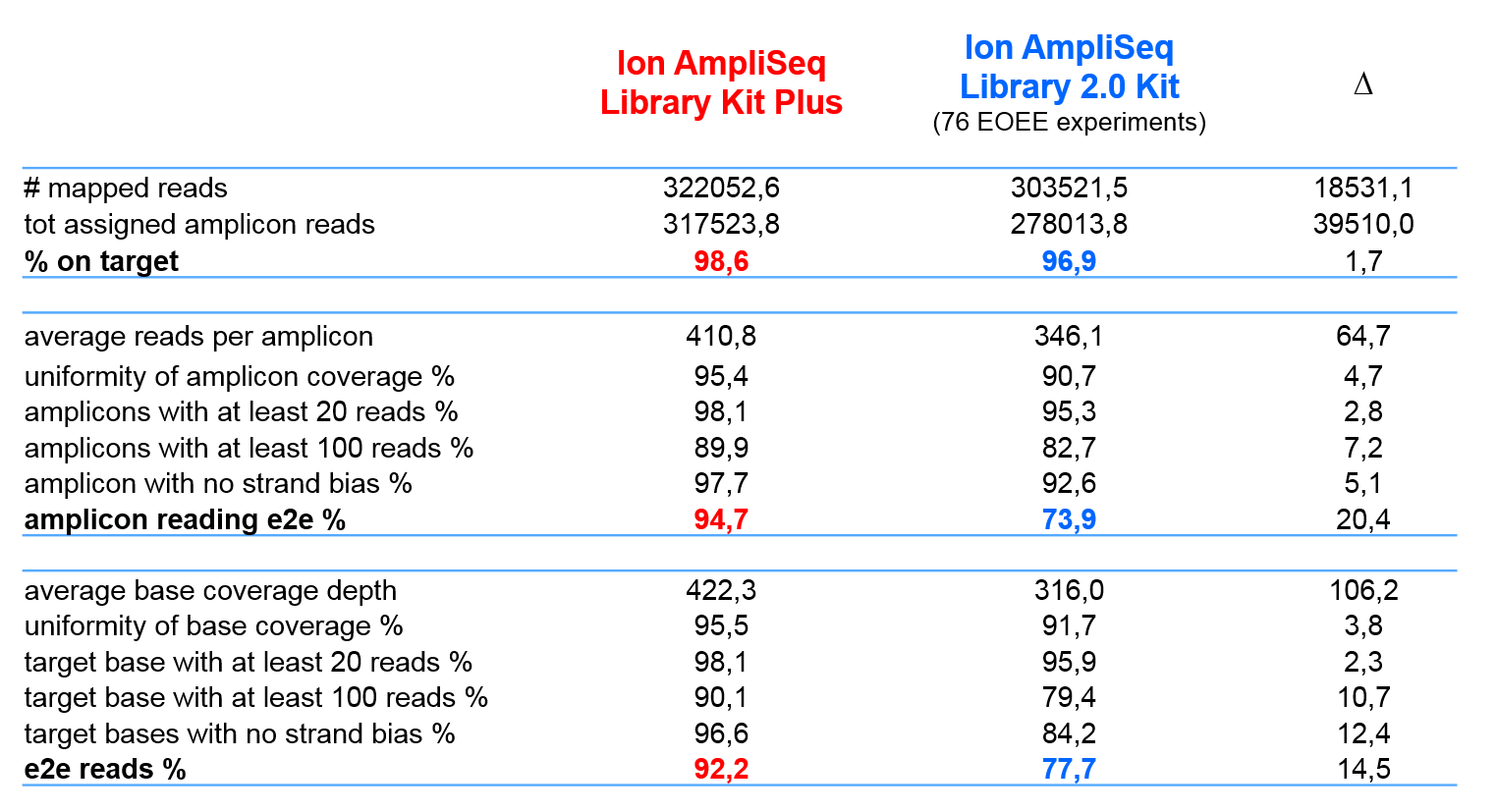
Figure 1. Coverage analysis of Ion AmpliSeq On-Demand panel using Ion AmpliSeq Library Kit Plus and Ion AmplISeq Library 2.0 Kit. Dr. Murgia’s lab evaluated coverage of the same On-Demand panel using the two library kits and observed improved performance with the new Ion AmpliSeq Library Kit Plus.
Ion AmpliSeq On-Demand panels can be built in Ion AmpliSeq Designer, a free online tool that enables researchers to configure NGS panels from a growing catalog of pre-tested genes across multiple research areas.
For more on Dr. Murgia’s research, and a demonstration of the capability of Ion AmpliSeq Designer, watch Dr. Murgia’s webinar “Application of targeted next-generation sequencing (NGS) panels in neurodevelopmental disorder research“.
For Research Use Only. Not for use in diagnostic procedures.




Leave a Reply