Touch DNA. Trace DNA. LCN DNA. It goes by a variety of names but they all conjure up the same concept- a sample quantity that is below recommend threshold in any stage of forensic DNA analysis workflows. How can forensic scientists get a DNA profile from such a minute sample? And why would you want to? (1) Back in 1997, van Oorschot and Jones reported getting a DNA fingerprint from a fingerprint, or perhaps better said, objects that had been handled (2). This expanded the number and types of evidence that could be investigated as well as the types of crimes – such as theft, armed robberies and assault.
Improvised explosive devices (IEDs) have seen widespread use since the early 20th century in intimidating activities toward military and non-military populations and communities world-wide. Advances in touch DNA analysis may now help to identify the suspects who are associated with making these weapons. The laboratory of Dr. Sheree Hughes-Stamm at Sam Houston State University (USA) has published a study on analysis techniques for human identification or ancestry determination on DNA samples recovered from pipe bombs that were spiked with known amounts of known DNA.
These test pipe bombs were made and spiked with > 800 cells (single source male Caucasian sample) in 5 spots on the end caps and the pipe shaft. The copper wires were stripped and spiked with 10 ul of blood from one of three individuals of differing ancestry (Asian, Caucasian or African American samples). There was no need to worry about safety as the local fire marshal’s department completed assembly and detonated the bombs in a secure location where evidence was collected and then brought back to the lab.
After DNA was extracted, samples were first evaluated using Applied Biosystems Quantifiler Trio DNA Quantification Kit which tests for total DNA quantity, presence of male DNA and the level of degradation. 44 of the 83 fragments showed detectable amounts of DNA and only 3 of those were shown to be severely degraded.
Then the samples were variously analyzed by three methods: STR analysis using Applied Biosystems GlobalFiler PCR Amplification Kit and analyzed on a capillary electrophoresis system, INNUL analysis from InnoGenomics Technologies, and SNP analysis for identity and ancestry determination using Applied Biosystems Precision ID NGS System.
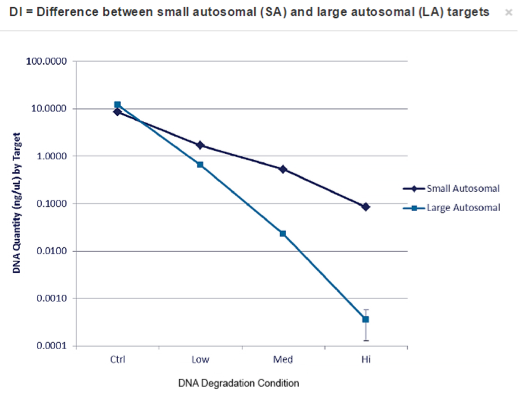
For STR analysis, 15 ul of neat sample was added to the GlobalFiler reaction where eight of the samples failed to amplify and two gave 100% recovery of all the alleles. But the 80% of these samples showed some allele drop-out, which comes as no surprise as they were all quite low in DNA input amount, with the higher amount of input showing somewhat lower drop-out rates. When analyzing which markers were affected, those in the higher size range showed degradation as was to be expected with these mildly degraded samples showing an average DI >6 from the Quantifiler Trio analysis.(3)
INNULs are retrotransposable elements that are dispersed throughout the human genome. They are bi-allelic markers with low mutation rates and can be detected via a small amplicon strategy. (4) In this study, INNULs generated successful genotyping with the degraded DNA samples using 16 ul of neat DNA. However, the RMP of this set of INNULs (1 x 10-8) is greater than STRs, and INNULs provided a lower RMP only when there were fewer than 14 STRs typed. (3) Given their small size, they could be a complimentary analysis method for highly degraded DNA samples.
 When using the Precision ID Identity Panel (124 SNPs) and next-gen sequencing, the input amount is only 6 ul of template, compared to 15 ul for STR analysis. This study used both 6 ul of input as well as 15 ul that had been concentrated to 6. The samples with just 6 ul contained 0.03-0.12 ng of DNA and yielded results with <50% allele calls but the concentrated samples with 0.4-0.27 ng provided for allele calls >70%. The larger number of SNPs in the panel provided for the lowest RMP of all the analyses.(3)
When using the Precision ID Identity Panel (124 SNPs) and next-gen sequencing, the input amount is only 6 ul of template, compared to 15 ul for STR analysis. This study used both 6 ul of input as well as 15 ul that had been concentrated to 6. The samples with just 6 ul contained 0.03-0.12 ng of DNA and yielded results with <50% allele calls but the concentrated samples with 0.4-0.27 ng provided for allele calls >70%. The larger number of SNPs in the panel provided for the lowest RMP of all the analyses.(3)
Lastly, six samples that were recovered from the copper wires were tested using 165 ancestry SNPs in the Precision ID Ancestry Panel. Five returned the correct ancestry though the confidence levels were a bit low. The input DNA of each sample was well below the suggested input of 1 ng which may explain the lower confidence levels.(3)
DNA recovered post-detonation from IEDs is expected to be of low quantity as well as low quality. But recent advancements in alternative marker types, though still in their early stages, show promise in helping to make identifications from these challenging sample types. And it is likely that more than one marker type will be needed to give the more complete picture as every sample and every case will have different needs.
To learn more about STR and NGS analysis methods for human identification, please visit thermofisher.com/hid.
- van Oorschot RA, Ballantyne KN, Mitchell RJ. Forensic trace DNA: a review.Investigative Genetics. 2010;1:14. doi:10.1186/2041-2223-1-14.
- van Oorschot RAH and Jones MK. DNA fingerprints from fingerprints.Nature. 1997;387:767. doi: 10.1038/42838.
- Tasker, Esiri et al. Analysis of DNA from post-blast pipe bomb fragments for identification and determination of ancestry. Forensic Science International: Genetics. 2017. Volume 28 , 195 – 202
- LaRue B, L, Sinha S, K, Montgomery A, H, Thompson R, Klaskala L, Ge J, King J, Turnbough M, Budowle B, INNULs: A Novel Design Amplification Strategy for Retrotransposable Elements for Studying Population Variation. Hum Hered 2012;74:27-35




Leave a Reply