
Extracting high-quality, intact genomic DNA is a foundational step for many downstream molecular applications—but not all DNA is created equal. High molecular weight (HMW) DNA extraction is critical for enabling advanced genomic analyses that depend on long, unfragmented DNA strands, especially in structural variant detection, genome assembly, and epigenetic profiling.
With the rise of long-read sequencing technologies, the ability to consistently isolate DNA fragments greater than 40 kb—and often exceeding 100 kb—is no longer a luxury but a requirement for cutting-edge genomic science.
Understanding the intricacies of DNA is crucial for advancements in genomics and biotechnology. Fundamental concepts such as genomic DNA, high molecular weight DNA, and the innovative techniques of long-read sequencing, have revolutionized the effect of de-novo sequencing by enabling the assembly of genomes without a reference. In this article, we will delve into the distinctions between genomic DNA structures, including high molecular weight DNA, and how they may affect long-read and short-read sequencing, shedding light on their unique applications and benefits.
Table of contents
- What is genomic DNA?
- What is high molecular weight DNA?
- Long read vs short read sequencing: what’s the difference?
- Comparing the advantages and applications of short-read and long-read sequencing
- What is the importance of HMW DNA for long read sequencing?
- HMW DNA extraction
- Understanding common HMW DNA extraction challenges
- FAQs for HMW DNA extraction
- More resources on HMW extraction and long read and short read sequencing
What is genomic DNA?
Genomic DNA (gDNA) refers to the entire set of DNA within an organism, including all its genes and non-coding regions. Genomic DNA can commonly be utilized in molecular applications such as PCR, sequencing, and next-generation sequencing.
What is high molecular weight DNA?
High molecular weight (HMW) DNA specifically refers to DNA molecules that are exceptionally long and intact, typically spanning hundreds of kilobases to megabases in length. While gDNA encompasses all the genetic material, HMW DNA is characterized by its large size and high integrity, making it particularly valuable for applications such as long-read sequencing and de-novo genome assembly, where long, continuous DNA strands are essential for accurate analysis and assembly.
Long read vs short read sequencing: what’s the difference?
Short-read and long-read approaches differ primarily in the length of DNA fragments they can analyze.
Short-read sequencing
Short-read sequencing — the most commonly-used form of massively parallel sequencing or next-generation sequencing (NGS) — is a widely adopted method that generates millions to billions of DNA fragments typically ranging from 50 to 300 base pairs in length. Its high throughput, low cost per base, and well-established informatics pipelines have made it the dominant sequencing technology in research and clinical labs over the past decade. From RNA-seq to targeted resequencing and microbial profiling, short-read platforms have powered countless life science breakthroughs. However, while short-read sequencing excels at variant calling and quantification, it struggles with repetitive regions, structural variants, and phasing—areas where long-read sequencing is increasingly favored. Short-read sequencing remains a popular approach due to its scalability, reliability, and broad application base.
Long-read sequencing
Long-read sequencing is a DNA sequencing technology that produces relatively long Mean Read Lengths, typically ranging from several thousand to over a hundred thousand base pairs in length. Long-read sequencing technologies are particularly valuable for applications such as de-novo genome assembly, structural variant detection, and resolving haplotype phasing, enabling a more comprehensive view of the genome.
What’s the difference?
Long-read sequencing is different than short read sequencing in that it sequences longer DNA fragments (thousands to hundreds of thousands of base pairs) compared to short-read sequencing, which utilizes shorter fragments (typically up to a few hundred base pairs), allowing for more accurate assembly and analysis of complex genomic regions. Short-read sequencing platforms, like next-generation sequencing technologies offered by Thermo Fisher Scientific Ion Torrent Chef/S5 systems or Thermo Fisher Scientific Ion Torrent Genexus Sequencers may provide the ability to sequence large numbers of short DNA fragments, which make it an excellent workflow integration with whole-genome sequencing and targeted sequencing. Table 1 below outlines some advantages of short-read and long-read sequencing technologies as related to several applications.
Comparing the advantages and applications of short-read and long-read sequencing
| Short-read | Long-read | |
|---|---|---|
| Advantages | High accuracy Cost effective High throughput Utilize standard genomic DNA | Longer reads May Improve SV detection compared to short read Resolves complex regions Utilize HMW genomic DNA (≥ 40 kb)* |
| Structural variations | Limited to small structural variations | Critical for large scale genomic alterations |
| De novo genome assembly | Less effective for new genome assemblies | Essential for assembling genomes from scratch |
| Haplotype phasing | Limited phasing capabilities | Determining phase over long distance |
| Epigenetic studies | Limited epigenetic analysis | Simultaneous genetic and epigenetic analysis |
| SNP and small indel deletion | Ideal for detection SNPs and small indels | Less effective for very small variants |
*DNA fragment size requirements vary widely based on application and platform
What is the importance of HMW DNA for long read sequencing?
High molecular weight genomic DNA (HMW gDNA) is used for long-read sequencing and other applications that require large DNA molecules, such as optical genome mapping. Due to the complexity of de novo sequencing and long-read sequencing applications requiring high molecular weight DNA, it’s vital to have high-quality isolated DNA greater than 40kb for most applications. In some instances, having greater than 100kb can facilitate even more optimal results. Navigating throughput needs, quality, and genomic size can pose difficulties in evaluation or lab generation.
HMW DNA extraction
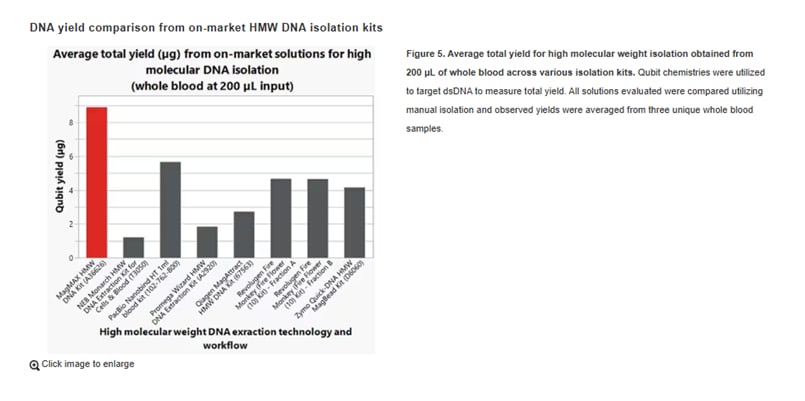
The Applied Biosystems™ MagMAX™ HMW DNA Kit was designed to obtain high molecular weight genomic DNA suitable for long-read sequencing platforms from manufacturers like Oxford Nanopore or PacBio. MagMAX HMW DNA kitwas optimized to obtain high quality yields and quality performance from fresh or frozen whole blood, cultured cells, and tissues.
With its magnetic bead-based technologies, the kit was optimized with workflows for manual isolation or automated using Thermo Fisher Scientific KingFisher™ purification instruments, enabling flexibility in sample throughput and lab automation needs.
Performance data for MagMAX HMA DNA kits are available online, including DNA yield, quality, fragment size, and fragment recovery by sample type.

Understanding common HMW DNA extraction challenges
Below are some examples of challenges that you might encounter when isolating HMW gDNA. We’ve included possible solutions to help achieve improved extraction quality, efficiency, and performance in downstream applications.
1. Low yielding samples
The MagMAX HMW DNA Kit is designed to yield a minimum of 3 µg of HMW gDNA from 200uL fresh/frozen whole blood, 1 X 106 cells and upto 8mg of tissue. A260/280 and 260/230 ratios greater than 1.8 are preferred to help ensure minimal protein and contaminant carryover.
Low yields and purity could be attributed to several causes across all sample types described further below.
Low yield across all sample types
Possible cause: The DNA sample was lost somewhere in the workflow.
Solution: If conducting a manual isolation, pipette tips may leave behind trace amounts of sample. Be sure to check the pipette tips before discarding. The HMW gDNA may get tangled in the tip during the washing steps when conducting isolation with manual workflows.
Possible cause: Insufficient mixing.
Solution: If large genomic DNA is observed to be tangled on the beads during the lysis/binding or washing step, be sure to break it up using a regular-bore pipette tip when utilizing manual workflows. Mixing the HMW gDNA-bead aggregate 5-10 times should be sufficient. Avoid over-mixing to prevent excessive shearing. Consider utilizing automated workflows on KingFisher purification instruments to remove user-error in mixing consistency. The KingFisher purification instrument has been verified to perform optimally with established mixing speeds as it is shown within the user instructions.
Possible cause: The beads were overdried.
Solution: If the beads appear cracked after the 2-minute drying at 50°C, try air-drying the beads at room temperature for 2 minutes when using manual workflows. Consider utilizing the optimized KingFisher purification protocols to remove user-error in drying consistency.
Possible cause: insufficient digestion of sample.
Solution: follow the recommended suggestions within the user-guide for the MagMAX HMW Kit for each specific sample digestion requirements. Avoid overloading the sample. Note that the Enhancer Solution (included in MagMAX HMW Kit) boosts Proteinase K activity but may also precipitate at lower temperatures. If the Enhancer Solution precipitates, ensure you incubate the solution at 37°C prior to use to ensure maximum efficiency. Consider isolating with a KingFisher purification instrument as the on-deck heating plate will help ensure consistent and sufficient heat to obtain proper digestion when utilizing the verified MagMAX HMW protocols.
Low yield in whole blood samples
Possible cause: Blood contains hemoglobin and other proteins which can be hard to wash away and properly digest
Solution: It is critical to transfer the sample to a new tube after the third wash to prevent carryover contaminants in the eluate with manual workflows. Consider utilizing automated workflows on KingFisher purification instruments which may reduce risk of sample-to-sample contamination introduced by user operator while helping ensure consistency of isolation from binding, washing, drying, and eluting the nucleic acid.
Possible cause: starting sample white blood cell (WBC) count may be low or starting samples may be clotted
Solution: A low WBC count means fewer cells are available for DNA extraction, leading to lower overall DNA yield. Blood clots can be difficult to isolate nucleic acid from, especially HMW genomic structures. Try to avoid clotted blood by maintaining best practices for storage conditions, blood stabilizers (like K2EDTA), and avoid over-mixing the sample before processing.
Low yield in cultured cell samples
Possible cause: isolating greater than 1 million cultured cells
Solution: The MagMAX HMW DNA Kit was optimized to process up to 1 million cultured cells. If attempting to process greater than 1 million cultured cells, consider increasing the reagent volumes in a linear ratio for all components. With manual workflows, ensure you utilize appropriate working solution containers which can fit the volume of reagents utilized. For automated KingFisher workflows, consider adjusting the KingFisher protocols utilizing the BindIt software (for KingFisher Flex purification system, KingFisher Duo Prime purification system) or BindIx software (for KingFisher Apex purification system) to reflect new working volume conditions. Keep in mind that the maximum volume allowance for a 96-mangetic head is 1mL while the maximum working volume for a 24-magnetic head is 5mL. No workflows beyond 1 million cultured cells or in a scaled up larger volume format have been verified by Thermo Fisher Scientific.
Low yield in tissue samples
Possible cause: Insufficient digestion may result in undigested tissue pieces being carried over into the washing steps.
Solution: Be sure to follow the digestion protocol listed in the user guide. Isolating from tissues exceeding the recommended mass may contribute to improper sample digestion. If isolating high molecular weight DNA manually, Invert the tubes 5-10x after adding the pre-digestion mixture to help ensure no tissue chunks are stuck in the cap prior to digestion. Check the tube mid-way through digestion to look for the presence of undigested tissue. Vortex if observed. Consider utilizing automated workflows on KingFisher purification instruments which may support a more cohesive digestion of the sample being used.
2. Viscous or brown eluent
Some bead carryover may be observed in the elution plate (automated extraction) or when the samples are transferred to new tubes (manual extraction). Some tissues, like spleen, are likely to contain significant amounts of DNA which could produce a more viscous or brown-colored eluent. Minor bead carryover into the elution is unlikely to interfere with downstream molecular applications.
If bead carryover is excessive or inhibitory to your downstream application:
Possible cause: The sample input is too high.
Solution: The workflow instructions for MagMAX HMW DNA Kit were optimized at ratios as claimed within the user manual. When utilizing less- or more- sample, those ratios may be affected and cause insufficient proportions especially as related to the surface area of the magnetic bead, which could cause bead carry-over in the elution. Follow the user guide to determine maximum inputs for each sample type. If your sample type is not listed, consider performing an initial titration experiment to determine the optimal input.
Possible cause: improper digestion
Solution: ensure that you are adding all components from the MagMAX HMW DNA Kit in the order specified within the userguide. If the order of operations is adjusted, the required vigorous mixing is not met, or the heat for digestion is not met then DNA eluate may appear “goopy,” thick, and pipetting may be difficult. If improper digestion is suspected, consider increasing the digestion time.
There are a few steps you can take to help reduce viscous eluent or bead-carryover.
- Use a magnetic stand to remove beads from elution.
Place the elution plate on a 96-well magnetic stand to ensure all magnetic beads are captured. Carefully transfer the clear supernatant to a new 96 deep-well plate or microcentrifuge tube. - Ensure complete digestion.
Ensure complete Proteinase K digestion to break down proteins effectively. If needed, perform additional off-board Proteinase K digestion as outlined in manual protocol. - Review user protocol instructions.
When working with high DNA-yielding tissues, follow the user guide recommendations for appropriate tissue input amounts.
3. HMW DNA degradation
One possible cause of HMW DNA degradation lies in the starting sample material.
Consider risks of overmixing, poor quality samples, complete sample digestion, or utilizing physical mechanical lysis which fragments the DNA recovered. Consider the following:
- Avoid mixing fresh whole blood samples with a serological pipette as this can create shearing of the DNA. Instead, gently invert the whole blood sample tube at least 5 times to ensure proper homogenization before transferring to the sample plate or tube.
- For frozen blood samples, limit to a single freeze/thaw cycle.
- For cells, ensure complete homogenization after adding the MagMAX Cell and Tissue Extraction Buffer.
- For tissue samples, do not use mechanical grinding. Instead, use gentle enzymatic digestion with Proteinase K.
- When isolating manually, carefully review the user guide for instructions on pipette tips (regular compared to wide-bore).
- Store eluates at appropriate temperatures (4ºC for short term and -20ºC for longer term storage when analysis will not be performed within 48 hours.
- Avoid freeze/thaw of the isolated nucleic acids.
FAQs for HMW DNA extraction
What are some of the applications of HMW DNA isolation workflows?
HMW DNA isolation has many downstream applications, including cancer genomics research involving the use of cell-free DNA (cfDNA) as biomarkers.
What sample types are compatible with the MagMAX HMW DNA kit?
The kit supports a range of inputs:
- Whole blood (fresh or frozen, ~200 µL)
- Cultured cells (~1×10^6 cells)
- Tissue (e.g. 2–8 mg)
Can this kit be used with automation systems?
Yes, MagMAX HMW DNA kits are compatible with KingFisher Duo Prime, Flex, and Apex systems. Automation can enable consistent, walk-away workflows in under two hours. Manual magnetic rack protocols are also available.
How many reactions are possible with the MagMAX HMW DNA kit?
Kit workflows support both manual and automated processing with up to 100 preps.
How does the MagMAX HMW DNA kit compare to others?
In direct comparisons, the MagMAX HMW kit achieved competitive or exceptional yields from whole blood versus others on the market, especially in blood volumes around 200 µL.
How long does the extraction take with a MagMAX HMW DNA extraction kit?
Extraction typically takes less than 2 hours for automated workflows. Manual protocols are also rapid and efficient.
More resources on HMW extraction and long read and short read sequencing
- Product Solutions: MagMAX HMW DNA extraction kits
- User guide for extraction kits
- Automation: KingFisher automated sample purification and extraction systems
- Demo request for automated processing systems
- Learn: Sequencing
##
© 2025 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.
For Research Use Only. Not for use in diagnostic procedures.






Leave a Reply