The Human Proteome Project (HPP) is an international project organized by the Human Proteome Organisation (HUPO). Its mission is to systematically map the entire human proteome by coordinating the efforts of many research laboratories around the world.1
The project is split into two divisions:
- The Chromosome-Centric Human Proteome Project
- The Biology/Disease Human Proteome Project
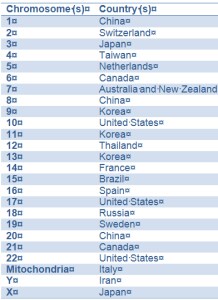 The Chromosome-Centric Human Proteome Project (C-HPP) was proposed by Young-Ki Paik1 and colleagues at several HUPO meetings from 2008 to 2011, the year the group officially formed. The essential idea behind the C-HPP is to organize emerging proteomic information by chromosome and gene. A 25-member international consortium will map and annotate the entire protein set coded by each human chromosome. Responsibility for maintaining the information for each of the 24 chromosomes has been assigned to different countries.2 The table below shows which country has claimed which chromosomes.
The Chromosome-Centric Human Proteome Project (C-HPP) was proposed by Young-Ki Paik1 and colleagues at several HUPO meetings from 2008 to 2011, the year the group officially formed. The essential idea behind the C-HPP is to organize emerging proteomic information by chromosome and gene. A 25-member international consortium will map and annotate the entire protein set coded by each human chromosome. Responsibility for maintaining the information for each of the 24 chromosomes has been assigned to different countries.2 The table below shows which country has claimed which chromosomes.
The C-HPP initiative does not alter the way researchers carry out proteomic experiments. Its primary function is to aggregate data sets from new experiments and five key existing resources: Ensembl v69, NeXtProt, Peptide Atlas, GPMdb and the Human Protein Atlas. C-HPP cross references these and measures findings against thresholds for high confidence protein identifications before making final entries into a Master Table. The initiative will also promote deep proteomic research and top-down protein variant analyses. As well, the project will stimulate development of new statistical and bioinformatic techniques to integrate genomic, proteomic and individual protein variation information.2
The team has estimated that approximately 65-70% of “expected” proteins have been identified with relative confidence.2 The notion of “expected” proteins is complicated by the fact that, in some cases, a protein is coded by multiple genes. Conversely, a single gene can code for multiple proteins. Nevertheless, a primary mission of the C-HPP is to concentrate research on finding “missing” or poorly characterized proteins. A substantial question in this regard is whether or not detection techniques require further refinement or whether the biology of the missing proteins thwarts their detection. For example, they may be present in extremely low quantities and still be active, or they may only be expressed at the embryonic stage of human development, or they may be so homologous to other proteins as to be indistinguishable.2
In addition to presenting the entire human proteome gene by gene, the C-HPP will catalogue protein variants generated through alternative splicing and coding SNPs, as well as a comprehensive list of major post-translational modifications.2
The value of this massive 10-year project will be borne out in its ability to uncover new drug targets, biomarkers for diagnosis and a comprehensive “parts” list of the protein isoforms that regulate major and minor cell signalling pathways.
References
2 Hancock, W., and Paik, Y. (2013) “A First Step Toward Completion of a Genome-Wide Characterization
of the Human Proteome,” Journal of Proteome Research 12 (pp. 1−5).




Leave a Reply