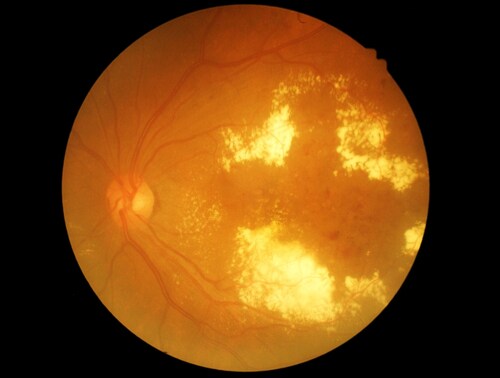
 Due to suboptimal awareness levels, newly diagnosed type 2 diabetic patients in the United States have often been diabetic for between four and seven years prior to diagnosis. Undiagnosed patients are therefore at risk of developing complications such as diabetic retinopathy, which can eventually lead to blindness, as a result of their unmanaged disease. Diabetic retinopathy is one of the most common and severe complications of type 2 diabetes. It has two main stages: non-proliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR). Chee et al. (2016) used proteomic studies of saliva to determine the changes in protein expression in the different stages of diabetic retinopathy.
Due to suboptimal awareness levels, newly diagnosed type 2 diabetic patients in the United States have often been diabetic for between four and seven years prior to diagnosis. Undiagnosed patients are therefore at risk of developing complications such as diabetic retinopathy, which can eventually lead to blindness, as a result of their unmanaged disease. Diabetic retinopathy is one of the most common and severe complications of type 2 diabetes. It has two main stages: non-proliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR). Chee et al. (2016) used proteomic studies of saliva to determine the changes in protein expression in the different stages of diabetic retinopathy.
The investigators collected saliva samples from 45 type 2 diabetes patients who visited the eye clinic at the University of Malaya Medical Centre between November 2013 and April 2014. They classified retinopathy severity according to the American Academy of Ophthalmology’s International Clinical Classification System for Diabetic Retinopathy and Diabetic Macular Edema, and then grouped patients according to clinical presentation:
- Type 2 diabetes without diabetic retinopathy (XDR), as control
- Type 2 diabetes with NPDR
- Type 2 diabetes with PDR
Chee et al. identified 315 proteins from the salivary proteome, using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific), which they annotated using gene ontology. The investigators assigned the proteins according to three classifications:
- Cellular component
- Biological process
- Molecular function
For quantitative analysis, they used cutoff values of less than 0.5 for fold change and greater than 2 to define differentially expressed proteins. As a result, they identified 119 differentially expressed proteins. A total of 117 salivary proteins were increased in the PDR disease group compared to the XDR disease group. They also found the the 82 salivary proteins that were increased in the PDR group were actually decreased in the NPDR group, while they did not find 34 of them in the NPDR group. Then the authors assigned the differentially expressed proteins to canonical pathways suggesting increased liver X receptor/retinoid X receptor (LXR/RXR) activation, farnesoid X receptor/retinoid X receptor (FXR/RXR) activation, acute phase response signaling, sucrose degradation V and actin-based motility regulation by Rho in the PDR disease group compared to the NPDR disease group.
Chee et al. suggest their findings indicate that the process whereby nonproliferative retinopathy progresses to proliferative retinopathy in type 2 diabetic patients is complex. Furthermore, their findings suggest that saliva is a feasible sample source for diabetic retinopathy biomarkers.
Reference
1. Chee, C.S., et al. (2016) “Association of potential salivary biomarkers with diabetic retinopathy and its severity in type-2 diabetes mellitus: a proteomic analysis by mass spectrometry,” PeerJ, 4(e2022), doi: 10.7717/peerj.2022.
Leave a Reply