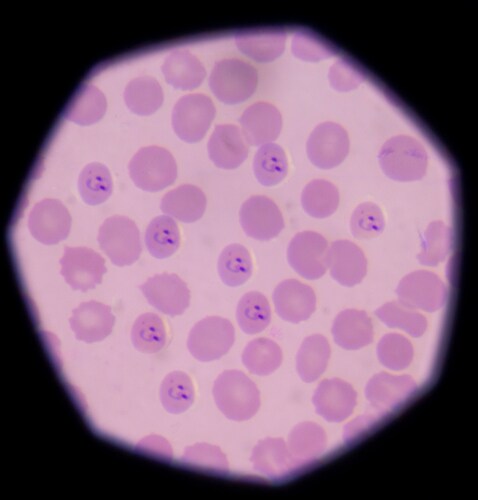
 Medical professionals frequently choose artemisinin-based combination therapy to treat uncomplicated Plasmodium falciparum malaria. One recommended combination treatment uses a pairing of dihydroartemisinin and piperaquine to effect high cure rates and exceptional tolerability in areas that demonstrate resistance to multiple drugs. A thorough understanding of piperaquine metabolism could assist in illuminating the compound’s pharmacological activity, clinical efficacy and toxicological profile.1
Medical professionals frequently choose artemisinin-based combination therapy to treat uncomplicated Plasmodium falciparum malaria. One recommended combination treatment uses a pairing of dihydroartemisinin and piperaquine to effect high cure rates and exceptional tolerability in areas that demonstrate resistance to multiple drugs. A thorough understanding of piperaquine metabolism could assist in illuminating the compound’s pharmacological activity, clinical efficacy and toxicological profile.1
Toward this end, Yang et al. (2016) show that liquid chromatography–high resolution mass spectrometry (LC-HRMS) plus multiple data-mining techniques in tandem can offer rapid metabolite profiling of drugs, including piperaquine.1 To do this, the team pretreated human (plasma, urine) and rat (plasma, bile, urine) samples by solid phase extraction before subjecting them to LC-HRMS using an LTQ-Orbitrap XL hybrid mass spectrometer and Xcalibur software (version 2.1) for data processing (both Thermo Scientific). They used tandem isotope pattern filter (IPF) and mass defect filter (MDF) post-acquisition data-mining techniques (MetWorks version 1.3 and Mass Frontier spectral interpretation software (version 6.0), Thermo Scientific).
The research team was able to rapidly identify six piperaquine metabolites (M1–M6) in human and rat samples. Of these, four (M1–M4) are major metabolites resulting from N-oxidation and/or carboxylation of the antimalarial compound. This study represents the first identification of metabolites M5 and M6 via a novel metabolic pathway for piperaquine: N-dealkylation. The team included proposed structural characterizations for the metabolites and pathway illustrations. The researchers note the absence of hydroxylated metabolites in the rat bile samples, which is a divergence from a previous study2 and likely due to unstable metabolites as a result of hydroxylation at the piperazine group.
In sum, Yang et al. offer LC-HR-Orbitrap-based MS as a solid analytical tool for rapid, comprehensive profiling of drug metabolites. The resultant data could impact current drug treatment protocols or lead to the discovery of novel therapeutic options.
Reference
1. Yang, A., et al. (2016) “Metabolite identification of the antimalarial piperaquine in vivo using liquid chromatography-high resolution mass spectrometry in combination with multiple data-mining tools in tandem,” doi: 10.1002/bmc.3689.
2. Tarning, J., et al. (2006) “Characterization of human urinary metabolites of the antimalarial piperaquine,” Drug Metabolism and Disposition 34(12) (pp. 2011–2019).
Post Author: Melissa J. Mayer. Melissa is a freelance writer who specializes in science journalism. She possesses passion for and experience in the fields of proteomics, cellular/molecular biology, microbiology, biochemistry, and immunology. Melissa is also bilingual (Spanish) and holds a teaching certificate with a biology endorsement.



Leave a Reply