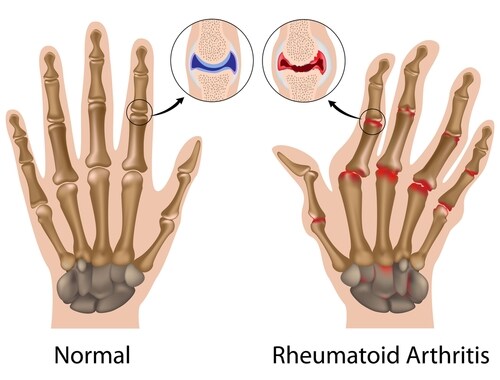
 Rheumatoid arthritis (RA) is a biomarker success story. With the development of current diagnostic biomarkers (anti-cyclic citrullinated peptide antibodies), clinicians have noted improvement in the diagnosis and early treatment of typical RA. Patients with atypical early RA, however, often experience delayed diagnosis and treatment, leading researchers to seek novel targets and a greater understanding of the pathophysiological process of the disease in all its forms.
Rheumatoid arthritis (RA) is a biomarker success story. With the development of current diagnostic biomarkers (anti-cyclic citrullinated peptide antibodies), clinicians have noted improvement in the diagnosis and early treatment of typical RA. Patients with atypical early RA, however, often experience delayed diagnosis and treatment, leading researchers to seek novel targets and a greater understanding of the pathophysiological process of the disease in all its forms.
With these goals in mind, Cheng et al. (2014) used high-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and tandem mass tagging (TMT) to screen differentially expressed RA-associated proteins.1 They used 60 individual serum samples (30 RA patients and 30 healthy controls), divided so that each of three groups contained samples from 10 patients with active early RA and 10 healthy controls. They enriched low-abundance proteins before subjecting the samples to SDS-PAGE, in-gel trypsin digestion, and isobaric labeling using TMT 126 for the healthy controls and TMT 129 for the RA samples (Thermo Scientific).
They desalted the labeled peptides and analyzed them with a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer interfaced with an UltiMate 3000 RSLCnano LC system (Thermo Scientific), with a 60-min gradient and a 0.30 μL/min flow rate. The Q Exactive mass spectrometer relied upon a fused silica capillary column with C18 resin (75 μm ID, 150 mm) and Xcalibur software (2.1.2, Thermo Scientific). The researchers performed one full-scan Orbitrap mass analyzer run (400–1800 m/z, 60,000 resolution) and 10 data-dependent MS/MS scans (27% normalized collision energy). They searched the spectra against the IPI human (v3.84) database with Proteome Discoverer software (Thermo Scientific) to determine relative protein quantification and identify differentially expressed proteins by fold change (fold ≥1.5).
The researchers identified 428 proteins, with a false discovery rate of ≤1%. Of these, in the RA samples, they determined that 26 proteins expressed abnormally. The 18 upregulated identifications included several proteins previously linked to RA (C-reactive protein isoform 1, SAA2-SAA2 protein, clusterin isoform 1, fibronectin, serum amyloid P component, complement factor H, glutathione peroxidase, and apolipoprotein E), as well as 3 over-expressed proteins that were novel discoveries: thrombospondin-1, ficolin-2, and an uncharacterized protein. The 8 downregulated proteins included proteins previously reported (alpha-2-macroglobulin, serum paraoxonase/arylesterase 1, and plasminogen) and newly identified under-expressed proteins: 58 kDa protein, isoform E of proteoglycan 4, cDNA FLJ54318, and inter-alpha (globulin) inhibitor H2.
Cheng et al. selected one protein of interest, upregulated ficolin-2 (FCN-2), using ELISA and an independent t-test to measure serum levels in RA patients and healthy controls. They found a concentration of 4.21±1.39 ng/mL in RA samples, which was significantly overexpressed from the 2.96±1.07 ng/mL found in control samples. The scientists also noted clinically relevant, positive correlations between the serum concentration of FCN-2 and swollen joint count (r=0.225, p<0.05), disease activity score in 28 joints (DAS 28; r=0.242, p<0.05), auto-antibody rheumatoid factor serum levels (r=0.243, p<0.05), and total Immunoglobulin M (IgM; r=0.270, p<0.05) in RA patients. They also observed clinically relevant correlations between DAS 28 (r=0.273, p<0.05) and IgM (r=0.359, p<0.01) in early RA patients.
The researchers posit that FCN-2 may be an acute-phase protein that mirrors reportable disease activity and participates in pathogenesis. They call for further functional evaluations of FCN-2, its relationship to RA, and its potential as a biomarker for early diagnosis.
Reference
1. Cheng, Y., et al. (2014, March) “Identification of Potential Serum Biomarkers for Rheumatoid Arthritis by High-Resolution Quantitative Proteomic Analysis,” Inflammation, doi: 10.1007/s10753-014-9871-8 [e-pub ahead of print]
Post Author: Melissa J. Mayer. Melissa is a freelance writer who specializes in science journalism. She possesses passion for and experience in the fields of proteomics, cellular/molecular biology, microbiology, biochemistry, and immunology. Melissa is also bilingual (Spanish) and holds a teaching certificate with a biology endorsement.



Leave a Reply