Go Digital PCR: Pt 4 – NGS Libraries is the fourth segment in our six-part series introducing you to digital PCR (dPCR) and now, how it enables highly precise next-generation sequencing (NGS) library quantification.
Simple, accurate, and affordable NGS library quantification with dPCR
dPCR is a simple and accurate technique for quantifying NGS libraries that helps avoid the potential miscalculation of library concentrations that may occur when using other techniques, such as microfluidic electrophoresis or spectrophotometry. dPCR uses library-specific assays that span both the forward and reverse adapters within a given library, measuring only those constructs that contain both library adapters.

CAPTION: Assay designs for NGS library quantification contain a forward primer, reverse primer, and probe.
Using dPCR to accurately quantify NGS libraries helps lower sequencing workflow costs by minimizing sample handling and eliminating the need to generate a standard curve or reference sample.
dPCR can also generate precise library concentrations in just a few hours following a simple workflow, thereby minimizing the need to re-run or repeat sequencing of samples. Library concentrations that are too high or too low lead to fewer total reads, lower sequencing yields, and reduced system throughput.
When to use dPCR to quantify NGS libraries
Standard libraries require an amplification step that enriches for correctly formed library constructs, which means that they have both the A and B adapters. This step minimizes the incorrectly formed constructs indicated by the single oval. An incorrect construct would mean they have only an A adapter or B adapter. As such, current detection methods, such as a spectrophotometer may be inadequate for quantifying this type of library construct.
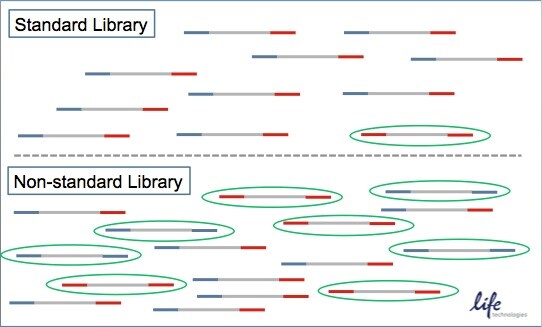
dPCR comes in handy for non-standard library protocols, which are typically PCR-free protocols, meaning that there will likely be a higher population of incorrectly formed library constructs as seen in the green ovals.
In non-standard libraries, amplification helps to enrich the correctly formed library constructs. Current quantification methods for quantifying NGS libraries include spectrophotometry and qPCR.
Spectrophotometry has long been used to quantify DNA and RNA. However, UV absorbance measurements are not selective and can’t distinguish between DNA, RNA, or protein. Values may also be affected by contaminants – free nucleotides, salts, and organics, e.g., ethanol and phenol, – and variations in base composition, which can lead to artificially high results. The sensitivity of spectrophotometry is often inadequate for quantifying DNA and RNA at low concentrations.
Real-time qPCR is a viable method for NGS library quantification, but requires a reference sample and the need to generate a standard curve. In high throughput laboratories sequencing different library types in parallel, the complexity of generating multiple standard curves can increase costs, time, and the potential for errors.
One significant potential error is PCR bias – differences in the amplification efficiency of templates or the inhibition of amplification by the most abundant templates in the late stages of amplification. Many protocols are leaning away from amplification to avoid PCR bias, which results in a smaller population of correctly formed constructs.
Specifically, spectrophotometer readings detect both correctly formed and incorrectly formed constructs, thereby potentially giving you an overestimation of library concentration.
The QuantStudio™ 3D Digital PCR System, together with TaqMan® assays for Ion Torrent™ and Illumina® library quantification, helps alleviate overestimation problems by detecting only correctly formed constructs to leave you with a precise library quantification. Accurate and precise NGS library quantification is a critical step in both Ion Torrent™ and other NGS workflows that helps maximize sequencing yields.
Precise quantification of Ion Torrent™ NGS libraries using QuantStudio™ 3D
The template preparation step in the NGS workflow is key to obtaining optimal sequencing yields on selected Ion Torrent™ platforms and is highly dependent on the library input amount – concentrations that are too high or too low can adversely affect total reads and system throughput.

CAPTION: NGS workflow using the QuantStudio™ 3D to accurately quantify NGS libraries prior to template preparation on the Ion OneTouch™ 2 System.
The QuantStudio™ 3D distributes a standard PCR reaction mix – diluted sample + QuantStudio™ 3D master mix + TaqMan® Assay – into a sealable chip containing 20,000 individual wells. Up-front sample dilution helps ensure that a portion of these partitions contain the target molecule, while other partitions do not, leading to positive and negative reactions. Following amplification on a dual flat block thermal cycler, the fraction of negative reactions is used to quantify the number of target molecules in the sample.
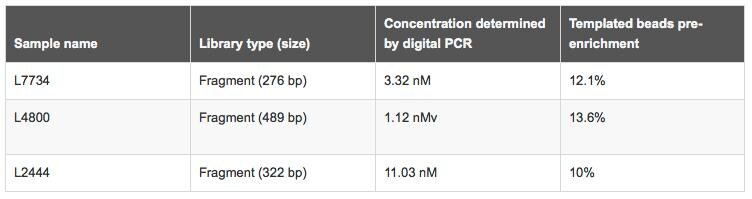
CAPTION: Correlation of QuantStudio™ 3D and Ion OneTouch™ 2 quality control data generated for four Ion Torrent™ libraries using flow cytometry prior to enrichment.
Absolute Quantification: Prior to determining final library concentration, the data must be adjusted based on the degree of 1) the sample dilution prior to preparing the reaction mix, and 2) the dilution that occurs when the reaction mix is formulated. QuantStudio™ 3D AnalysisSuite™ Cloud Software is available to facilitate this process.
Correlation: Dilution of the library based on the calculated concentration (nM) is necessary for template bead preparation using the Ion OneTouch™ 2 System. This is followed by a quality control step that uses the Qubit™ 3.0 Fluorometer to assess the quality of the templated beads generated in a sample prior to enrichment. Optimal library quantification corresponds to a library dilution that results in 10%-30% of beads containing amplified template. Samples within the recommended range generally deliver higher quality data.
Conclusion
Achieving optimal NGS yields requires high-quality template preparation. Using dPCR to quantify libraries helps decrease overall sequencing costs by enabling sequence quality that might otherwise result in additional downstream work or require duplication of steps. The QuantStudio™ 3D, together with library-specific TaqMan® Assays, provides an affordable and easy-to-use dPCR methodology that enables highly precise template generation, with minimal sample handling and without need to generate a standard curve or reference sample.
Learn More About QuantStudio™ 3D Digital PCR and NGS Libraries
Learn more about how the QuantStudio™ 3D Digital PCR System provides precision quantification of NGS libraries with minimal sample handling and without the need to generate a standard curve. For more on library offerings we suggest this application note: Precise quantification of Ion Torrent™ NGS libraries using QuantStudio™ 3D and this video: Quantifying Next-Gen Sequencing Libraries using digital PCR on the QuantStudio 3D with Peter Schweitzer, PhD, Genomics Facility, Cornell University. The NGS portion of the talk picks up at 4:10 into the video.
Finally, take a break from your busy day and log in to this informative on-demand webinar, Beyond the Limits with Digital PCR,
with featured speaker Dr. Lovorka Degoricija, Staff Scientist, Digital PCR Technology, Thermo Fisher Scientific. Learn how digital PCR on the QuantStudio™ 3D Digital PCR System can improve your Ion Torrent™ and other NGS sequencing workflow.
Read all of the Let’s Go Digital PCR Series:
Go Digital PCR: Pt 1 – Next-Gen Quantification
Go Digital PCR: Pt 2 – QuantStudio™ 3D
Go Digital PCR: Pt 3 – Copy Number Variation
Go Digital PCR: Pt 4 – NGS Libraries
Go Digital PCR: Pt 5 – Rare Mutations
Go Digital PCR: Pt 6 – Emerging Analyses



Leave a Reply