Go Digital PCR: Pt 5 – Rare Mutations is the fifth segment in our six-part series introducing you to digital PCR (dPCR) and how it is capable of detecting and quantifying rare mutations for low-prevalence targets in cancer research samples.
Rare mutation detection is a powerful tool in cancer research
In oncologic research, rare mutation detection among a large population of wild type cells poses a significant challenge. The accumulation of mutations in oncogenes or tumor suppressor genes is an important aspect of tumor formation. Acquisition of these mutations in a tiny subset of somatic cells can be sufficient for cancer initiation or progression.
Common SNP genotyping technologies, such as capillary electrophoresis or real-time qPCR, are effective at detecting mutant cells with prevalence no lower than about 20% – approximately one in five cells. But because these mutations are usually present only in a very small number of cells, they require an assay that delivers high signal-to-noise and low false-positive rates. dPCR has that capability. By subdividing a sample into thousands of individual PCR replicates, the total number of molecules in any given reaction is greatly reduced, effectively enriching for the sequences of interest and diluting out the wild type background.
There are also times where dPCR is useful in detecting very low quantities of nucleic acids in heterogeneous samples, i.e., allele detection. In tumor samples, for example, dPCR can be used to determine whether a small percentage of cells have a mutant allele within an oncogene.
Researchers are now able to detect low-frequency mutations by combining TaqMan™ fluorogenic 5’ nuclease chemistry with dPCR methodology using the QuantStudio™ 3D Digital PCR System. Together with TaqMan® SNP Genotyping Assays, the resulting QuantStudio™ 3D rare mutation analysis solution helps achieve the high levels of performance required to detect and absolutely quantify mutations present at a very low frequency.
Enables rare mutation detection in just hours with the QuantStudio™ 3D Digital PCR System
The QuantStudio™ 3D Digital PCR System is a simple and affordable dPCR platform for enabling the detection and quantification of rare mutations for low-prevalence targets in cancer research samples. The QuantStudio™ 3D leverages high-density nanofluidic chip technology to partition a sample into as many as 20,000 independent reaction wells. Comprised of consistently sized wells etched in a solid silicon substrate, the QuantStudio™ 3D Digital PCR Chip enables straightforward and robust sample partitioning that allows thousands of data points to be analyzed per run. The highly controlled sample loading and low reaction dropout combine to ensure higher precision for absolute quantification.
Key advantages of using the QuantStudio™ 3D rare mutation analysis solution include:
• Enables optimized dPCR performance
• 38 wet lab-validated TaqMan® SNP Genotyping Assays targeting 60 somatic mutations
• Helps detect and quantify rare mutant prevalence as low as 0.1%
• Single-tube format includes wild type and mutant alleles, with small and large reaction options
• Advanced algorithms help streamline quantification of rare mutations
Greater precision with wet lab-validated TaqMan™ SNP Genotyping Assays
Dr. Iain Russell, Sr. Product Manager for TaqMan® Assays, spoke of the most recent QuantStudio™ 3D enhancements to benefit rare mutation analysis in an interview at the American Society for Human Genetics (ASHG) 2014 annual meeting. Specifically, Dr. Russell described the QuantStudio™ 3D rare mutation analysis solution’s subset of 38 wet lab–validated TaqMan® SNP Genotyping Assays for use in detecting and quantifying rare somatic mutations, e.g., EGFR, BRAF, KRAS, PIK3CA, and JAK2. The assays are wet lab-validated specifically for our QuantStudio™ 3D Digital PCR System and allow you to quantify rare mutants at a prevalence as low as 0.1%.

CAPTION: This is an example of dPCR quantification of a rare KRAS allele in which a KRAS G12V mutation was detected. Each sample represents a fraction of mutant plasmid to wild-type genomic DNA. The 0.1% target represents mutant genomic DNA from a mutant cell line in a wild-type genomic DNA background. All samples were analyzed using QuantStudio™ 3D AnalysisSuite™ Cloud Software with the relative quantification module. (A)Targeted mutation rate: 10%. Mutation detection rate: 10.384% with a confidence interval of 8.235–13.076%. (B) Targeted mutation rate: 1%. Mutation detection rate: 1.393% with a confidence interval of 0.772–2.507%. (C) Targeted mutation rate: 0.1%. Mutation detection rate: 0.142% with a confidence interval of 0.064–0.310%. (D) Wild type control sample.
Target selection for the wet lab–validated SNP genotyping assays was based on frequency of occurrence as well as targets that exist on the Ion AmpliSeq™ Cancer Hotspot Panel. The assays are single-tube assays that contain primers and a probe for both wild type and mutant alleles, and are available in 12- and 450-reaction sizes.
Helps you perform absolute quantification with confidence
QuantStudio™ 3D AnalysisSuite™ Cloud Software, allows users to quantify the percentage of mutation in their samples from the data collected on the QuantStudio™ 3D Digital PCR System.
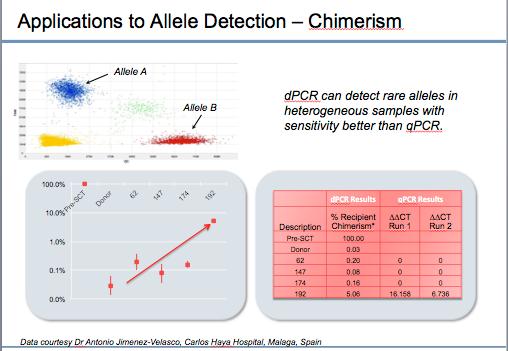
In applications involving rare allele detection, AnalysisSuite™ Software sequesters the results of individual PCR reactions into four different quadrants. The upper left quadrant (blue dots) is almost exclusively allele A, while the lower right quadrant is almost exclusively allele B (red dots). The green spots are the wells with both allele A and B, and the yellow dots are unamplified.
* These %s were calculated by Excel using data from AnalysisSuite™ Software
To illustrate this a collaborator Dr. Antonio Jimenez-Velasco (Carlos Haya Hospital, Malaga Spain) sent Thermo Fisher Scientific researchers leukemia samples post bone marrow transplant. Assays were selected where the bone marrow donors possessed a null allele of a locus and the bone marrow recipients samples possessed an insertion allele in the same locus, facilitating discrimination. Blood samples were collected from the donors and recipients before transplant and various times after transplant. The presence of the two loci in the samples was quantified by dPCR.
The graph shows the results from one recipient sample. Before the bone marrow transplant – Pre-SCT – the genotype of the circulating cells was 100% of the host’s genotype. The donor, shown in the next position had about 0.1% of the recipient’s allele. Over time, you can see that after 62 days, 147 days, and 174 days, there are low levels of the recipient sample’s original genotype. However, it starts to come up after 192 days, indicating that the recipient sample’s cells were repopulating the bone marrow. Error bars represent a calculated 95% confidence interval.
The collaborator was still unable to detect the chimerism before 192 days by traditional PCR. dPCR was more sensitive and picked up the chimerism (reappearance of original allele) earlier than traditional PCR.
Note: Mixed chimerism occurs in a leukemia sample when both the recipient and donor cell samples are present in bone marrow. It’s routinely analyzed after stem cell transplantation.
Conclusion
I hope this primer on dPCR and rare mutation detection and quantification has been enlightening and that the links herein will help your understanding of dPCR even more. To recap, the QuantStudio™ 3D rare mutation analysis solution offers your lab:
• Optimized digital PCR performance enhanced by 38 wet lab–validated TaqMan® SNP Genotyping Assays targeting 60 somatic mutations
• High sensitivity to detect and quantify rare mutation prevalence as low as 0.1%
• Cost-effective and convenient single-tube format that includes wild type and mutant alleles with small and large reaction options
• Streamlined analyses with advanced algorithms for better quantification of rare mutations
Learn more about dPCR and rare mutation detection and quantification
Learn more about how the QuantStudio™ 3D Digital PCR System enables detection and quantification of rare mutations for low-prevalence targets in cancer research samples. For more on the latest TaqMan® SNP Genotyping Assays available for rare mutation detection, take a moment to watch this recent video interview with Dr. Iain Russell, Life Technologies’ Sr. Product Manager for Digital PCR Technology at ASHG 2014: Rare Mutation Detection w/Digital PCR
In the world of webinars, we offer you several ways to expand your awareness of dPCR and rare mutation detection and quantification.
First up is Expanding the PCR Tool Box with Digital PCR: A review of absolute quantification and mutation detection applications using the QuantStudio™ 3D Digital PCR System. Steve Jackson, PhD, Thermo Fisher Scientific’s Associate Director of Product Applications in the Genetic Analysis division is the speaker. You’ll hear how digital PCR expands the application boundaries of traditional real-time PCR and see real world examples of how you can go beyond measuring Ct to detecting individual molecules, including oncogenic alleles in FFPE cancer specimens, and gaining additional sensitivity and precision for a variety of experiments.
Next, if you’re entirely new to dPCR, Dr. Iain Russell, Sr. Life Technologies’ Product Manager for Digital PCR Technology, can help you wrap your head around the basics with his webinar: Introduction to Digital PCR – Sensitive, precise, absolute quantification with chip-based digital PCR Listen in as he covers the benefits of digital PCR, how it works, and how to use it for applications that require increased sensitivity and precision. You’ll also learn how the QuantStudio™ 3D Digital PCR System employs a chip-based technology to perform simple and robust digital PCR.
Read all of the Let’s Go Digital PCR Series:
Go Digital PCR: Pt 1 – Next-Gen Quantification
Go Digital PCR: Pt 2 – QuantStudio™ 3D
Go Digital PCR: Pt 3 – Copy Number Variation
Go Digital PCR: Pt 4 – NGS Libraries



Leave a Reply