Search
The Max Planck Institute of Neurobiology uses Amira software for axon tracing from head to toe
Customer Spotlight
Ali Ertürk and his collaborators described a novel histo-chemical technique to clear spinal cord tissue of adult mice for fluorescence microscopy imaging of the intact spinal cord.
Previously, the high lipid content of the spinal cord tissue in adult mice allowed imaging of the spinal cord only through destructive histological methods, making the tracing of complete axons impossible. As applied in this study, the improved histo-chemistry enabled Ertürk and his colleagues to show that not only growth-competent axons, but also axons previously considered growth-incompetent show regenerative features after spinal cord trauma.
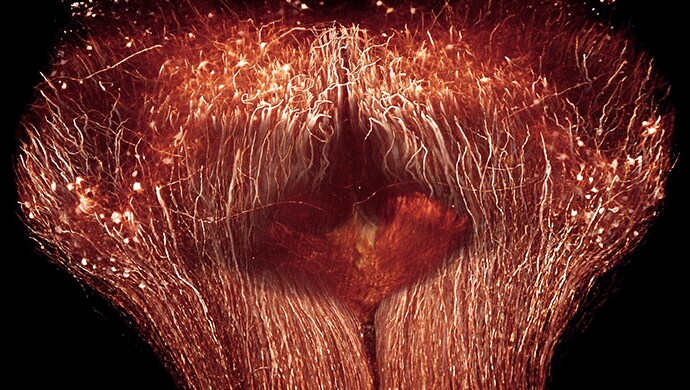
As a long term Amira software user, Ertürk trusted the system , with its filament tracing and visualization features, to be the tool of choice. Using Amira software's skeletonization modules and filament editing tools, he succeeded in tracing axons from the spinal cord lesion site to their dorsal root ganglions, thereby validating that they were derived from neurons whose peripheral axon had been cut. The tracing of axons up to their tip shows the great potential of 3D imaging to clearly distinguish regenerated axons from unharmed axons (Figure 1), all at a resolution of traditional histology methods.
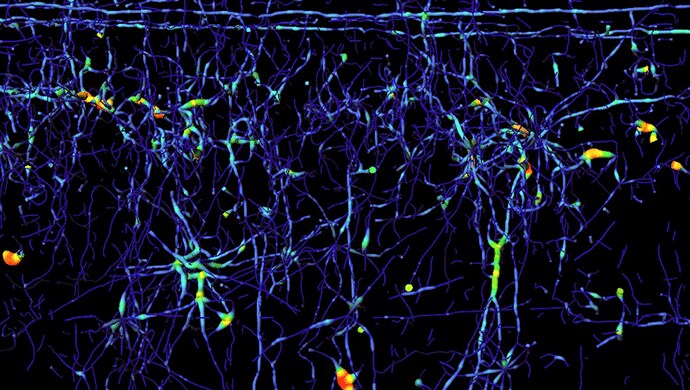
For the visualization of the 2-photon microscopic images Ertürk relied on Amira software's shaded volume rendering (see Figures 1 and 2). Together with its corner cut functionality (Figure 2) this not only generates beautiful illustrations of the massive parallel wiring inside the spinal cord but also allows one to virtually dissect the tissue and gain information about its spatial organization. The traced axon trajectories were instructively visualized using Amira software's tube rendering module. Figure 3 shows the network of fibers rendered as tubes colorized by their diameter.

Amira software's flexibility and support for other image processing platforms such as Matlab allowed Ertürk to easily exchange the tracing results with 3rd party applications, which he used to develop custom quantification methods.
As a last step, 3D videos were created using Amira software's DemoDirector and MovieMaker modules, which provide imitless freedom for creating 3D animations.
Ultimately, the 3D imaging methods will enable researchers to establish the precise wiring of the entire nervous system.
The Max Planck Institute of Neurobiology (MPIN) is situated in Martinsried, at the southwest border of Munich. The institute is devoted to basic research and investigates the basic functions, structure and development of the brain and the nervous system.
About Amira & Avizo 3D Software
Amira and Avizo are high-performance 3D software for visualizing, analyzing, and understanding scientific and industrial data coming from all types of sources and modalities.
Authors/Researchers: Ali Ertürk1,2, Christoph P Mauch3, Farida Hellal1,4, Friedrich Förstner5, Tara Keck6,7, Klaus Becker8,9, Nina Jährling8-10, Heinz Steffens11, Melanie Richter12, Mark Hübener6, Edgar Kramer12, Frank Kirchhoff11,13, Hans Ulrich Dodt8,9 & Frank Bradke1,4
Affiliations:
1 Max Planck Institute of Neurobiology, Axonal Growth and Regeneration, Martinsried, Germany.
2 Department of Neuroscience, Genentech, South San Francisco, California, USA.
3 Max Planck Institute of Psychiatry, Munich, Germany.
4 Deutsches Zentrum für Neurodegenerative Erkrankungen, Axonal Growth and Regeneration, Bonn, Germany.
5 Max Planck Institute of Neurobiology, Systems and Computational Neurobiology, Martinsried, Germany.
6 Max Planck Institute of Neurobiology, Cellular and Systems Neurobiology, Martinsried, Germany.
7 Medical Research Council Centre for Developmental Neurobiology, King's College London, Guy's Hospital Campus, London, UK.
8 Technical University Vienna, Institute of Solid State Electronics, Department of Bioelectronics, Vienna, Austria.
9 Center for Brain Research, Medical University of Vienna, Section of Bioelectronics, Vienna, Austria.
10 University of Oldenburg, Department of Neurobiology, Oldenburg, Germany.
11 Glial Physiology and Imaging, Department of Neurogenetics, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
12 Development and Maintenance of the Nervous System, Centre for Molecular Neurobiology, Hamburg, Germany.
13 Molecular Physiology, Institute of Physiology, University of Saarland, Homburg/Saar, Germany.
Images and text are courtesy of Ali Ertürk, Max Planck Institute of Neurobiology