Search
OncoScan CNV Assays for Analysis of FFPE Tumor Samples
Accurate analysis of copy number changes
The genomes of cancer cells often contain changes in their chromosomal architecture and such alterations can be critical to tumor formation and growth progression. Understanding the copy number makeup of solid tumors provides valuable insights into tumor biology, evolution, and resistance to therapy. Studying chromosomal abnormalities and genomic instability is therefore key to fully profiling solid tumors and identifying potentially prognostic and predictive biomarkers.
Catch critical copy number changes with OncoScan CNV Assays. The OncoScan CNV Assay and OncoScan CNV Plus Assay (previously known as OncoScan FFPE Assay Kit) enable accurate analysis of copy number changes and allelic imbalances across the entire genome. These OncoScan CNV assays have been shown to perform well with highly degraded DNA, such as that derived from FFPE tumor samples of various ages, and with low amounts of DNA starting material, making the assay a natural choice for clinical cancer research.
Whitepaper: OncoScan CNV and CNV Plus Assays: comprehensive solutions for whole-genome copy number analysis of FFPE tumor samples
Whole-genome copy number analysis is key to clearly identifying biomarkers that are associated with response or resistance to therapy, are known to have prognostic value, or can distinguish between aggressive and nonaggressive disease states. In this white paper, learn how molecular karyotyping analysis by high-resolution microarray complements, and may one day replace, traditional cytogenetic techniques.
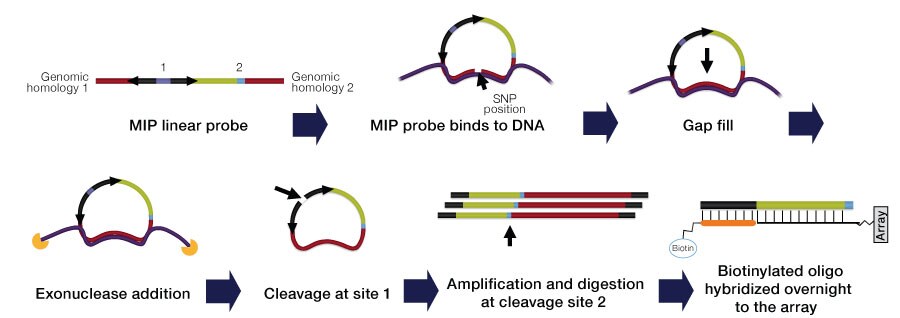
The OncoScan CNV assays utilize molecular inversion probe (MIP) technology (Figure 1), proven for identifying copy number alterations, loss of heterozygosity (LOH), copy neutral LOH (cnLOH), and somatic mutations.
Available in two versions
- OncoScan CNV Assay—high-density copy number coverage across 900 cancer research genes and standard coverage across the whole genome
- OncoScan CNV Plus Assay—same copy number coverage as the OncoScan CNV Assay plus a somatic mutation panel covering 64 mutations in 9 genes
OncoScan CNV assay features
- Whole-genome copy number analysis—detect structural variants such as deletions, duplications, and unbalanced translocations that are not well characterized by short-read or targeted sequencing
- Comprehensive coverage—whole-genome analysis of genes with established significance as well as those with emerging evidence, thus helping to reduce future re-verification burden
- Low sample input—get results in 72 hours from only 80 ng of FFPE-derived DNA
- All-in-one assay—detect chromosomal arm aberrations, focal changes, LOH, and cnLOH in a single assay, helping reduce cost and processing time
- Fast turnaround time—go from sample to answer in just three days
Simple whole-genome data analysis and reporting
Chromosomal Analysis Suite (ChAS) is a simple yet powerful analysis software enabling you to view and summarize genome-wide chromosomal aberration data from OncoScan CNV assays with just a few clicks.

On-demand webinar: Exploring the genetic landscape of solid tumors using whole-genome copy number analysis
This webinar explores the importance of cytogenetic analysis for solid tumor profiling, specifically in the research of leiomyosarcoma (LMS) and melanoma.
- Learn about the importance of copy number analysis in solid tumor research and its relevance for identifying biomarkers
- See how OncoScan CNV assays can be used identify relevant and informative copy number changes in LMS and melanoma
Resources
For Research Use Only. Not for use in diagnostic procedures.
