Search Thermo Fisher Scientific
Label Transfer Protein Interaction Analysis
Label transfer methodology
In a typical label transfer reaction, a bait protein reacts with a reactive moiety of a label transfer reagent (LTR), forming a covalent bond with the heterobifunctional crosslinker linked to a radioactive, fluorescent or biotin label, as diagramed below. This labeled bait protein is then allowed to interact with prey protein in vitro to form a complex, after which the reaction is exposed to UV light to activate a photoreactive group moiety on the crosslinker, which then covalently binds to the prey protein. The label transfer is completed by cleaving the crosslinker spacer arm to release the bait protein, leaving the label attached to the interacting prey protein, which can be detected through multiple methods, including western blot analysis, protein sequence analysis and mass spectrometry. A biotin label is especially useful in this process because it can be used for both the purification and detection of the prey protein.
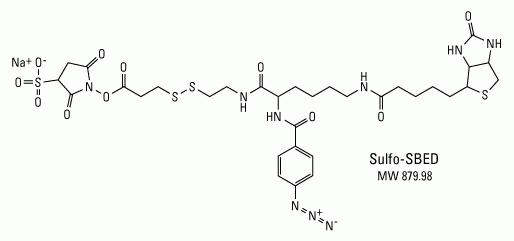
Label transfer chemistry.Sulfo-SBED has an amine-reactive NHS-ester (left side), aryl azide, photoreactive group (center with nitrogens) and biotin (right side). Transfer of the biotin from the amine on the bait protein to the target molecule occurs through reduction of the disulfide bond between the NHS-ester and photoreactive group.

General diagram of a label transfer reaction.
Bioconjugation and crosslinking technical handbook
Learn how to optimize your bioconjugation strategies with our updated Bioconjugation and crosslinking technical handbook. This easy-to-use guide overviews our portfolio of reagents for bioconjugation, crosslinking, biotinylation, and modification of proteins and peptides.
Achieve the most efficient modification for your typical applications including:
- Protein and peptide biotinylation
- Antibody labeling with fluorophores and biotin
- Immobilizing biomolecules to surfaces
- Capturing protein interactions
Traditional label transfer reagents
Iodinated labels for autoradiography
The earliest examples of label transfer reagents incorporated a photoreactive phenyl azide group that contained a hydroxy-phenyl modification on the ring. The phenolic hydroxyl activates the ring for substitution reactions to occur ortho or para to its position. These compounds can be radio-iodinated using typical oxidation reagents such as chloramine T or a commercial iodination reagent. Iodination of the crosslinker with I-125 prior to its use results in a radioactive label transfer reagent that can tag an unknown interacting protein with a radiolabel after cleavage of the crosslinker's spacer arm.
Iodination beads. The coated Thermo Scientific Pierce Iodination Beads provide for efficient iodine labeling and help to prevent damaging oxidative effects normally associated with chloramine T and other solution-based methods.
The crosslinker is first radio-iodinated and then reacted with a bait protein, typically through available amine groups. This modified protein is then added to a sample and allowed to interact with other proteins, and the sample is exposed to UV light to photo-crosslink the interacting complex. At this point, the label facilitates the detection of the interacting proteins, or the complex can be cleaved and the radiolabel transferred to the protein interacting with the bait. The now radiolabeled, unknown protein is then detected by autoradiography after separation by electrophoresis and western blot analysis.
The first reagents employed using this method were bifunctional and were designed so that the photoreactive moiety bears the transferable label. These molecules are either amine or sulfhydryl reactive and are labeled radioisotopically with I-125.
Disadvantages of traditional iodinated reagents
Although these reagents have been used successfully to obtain data on protein interactions, they possess some inherent deficiencies compared to trifunctional reagents designed for label transfer applications. The user should be aware of the following characteristics of these reagents:
- Reagents require labeling with I-125 before use, and the efficiency of label incorporation is low.
- The photoactivation step can result in several unproductive pathways that lower the crosslinking yield between bait and prey.
- The I-125 label can be released during the light reaction, causing nonspecific labeling of the protein(s) in the mix.
- Some molecules lack appropriate groups that can be iodinated, and thus may require modification at primary amines using Bolton-Hunter Reagent.
Newer kinds of compounds have been developed as trifunctional reagents that more adequately segregate the reactive sites from the label. Increasingly, these new kinds of reagents include non-radioisotopic labels such as biotin.
Nonradioactive label transfer reagents
Fluorescence
Subsequent designs of bifunctional label transfer reagents use nonradioactive labels to avoid the safety issues posed by I-125. Fluorescent constituents designed into cleavable photoreactive crosslinkers provide a method to transfer a fluorescent label to an unknown interacting protein. Using this approach, the reagent is non-fluorescent prior to exposure to UV light, but upon photolyzing and coupling to interacting proteins, the crosslinking molecule fluoresces.
These reagents also have a disulfide bond that can be reduced, resulting in the cleavage of the crosslinked proteins and the transfer of the label to the unknown interacting species. The fluorescently labeled interacting protein can then be followed in cells to determine the site of interactions or the fate of the proteins after the interaction occurs.
Because fluorescent compounds are inherently unstable when exposed to light for extended purposes, this makes them less than ideal as label transfer reagents. For researchers wishing to avoid radioactivity, we recommend the use of a label transfer reagent that includes a biotin tag rather than a fluorescently labeled reagent.
Biotin
Label transfer reagents can also have biotin built into the bifunctional structure of a crosslinking molecule. After reacting with the bait and prey proteins as diagramed above in the introductory section, the crosslinker is cleaved, resulting in a biotin-labeled prey protein that can be used to detect the protein.
This type of trifunctional label transfer method differs substantially from the bifunctional methods previously discussed because of the wide variety of methods available to detect the prey protein. Because the protein is biotinylated, both chromogenic and fluorescent labeling approaches can be used to detect the protein via avidin-, streptavidin- or Thermo Scientific NeutrAvidin Protein–conjugated horseradish peroxidase (HRP), alkaline phosphatase (AP) or a fluorescent molecule. Additionally, the biotin handle can be used to purify the prey protein or protein fragments using avidin-, streptavidin- or NeutrAvidin protein–conjugated agarose or magnetic bead chromatography.
There are numerous defining structural and functional properties of Sulfo-SBED, including:
- Amine-reactive NHS-ester group—for labeling a purified "bait" protein at the N-terminus and side chain of lysine residues
- UV light-activatable aryl azide group—for crosslinking nonspecifically to the protein side chains and backbone of the interacting protein after allowing protein binding to occur
- Cleavable disulfide bond (S-S)—can be reduced to release the crosslinker from the original "bait" protein
- Biotin group—remains attached to target interacting protein after cleaving the disulfide bond, thereby tagging the previously unknown interacting protein(s) for affinity purification and detection
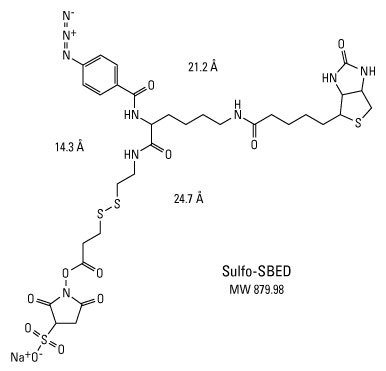
Structural and functional properties of Sulfo-SBED for biotin label transfer.
Since biotin was first used in label transfer applications, this approach is increasingly used to detect protein–protein interactions, as listed below:
- Define the interactions of complexes with activator domains
- Clarify the mechanism of protein complex assembly
- Study docking site and factor requirements for binding
- Describe the binding contacts of interactors
- Confirm the recognition of a specific phosphoepitope
- Search for putative binding partners
- Gain insight into chaperone-mediated refolding interactions
- Investigate the mechanism of protein interaction
- Facilitate receptor activity-directed affinity tagging (re-tagging)
- Detect low-abundance protein receptors
- Understand drug–receptor interactions
Recommended reading
- Neely KE et al. (2002) Transcription activator interactions with multiple swi/snf subunits. Mol Cell Biol 22:1615–25.
- Ishmael FT et al. (2002) Assembly of the bacteriophage t4 helicase: Architecture and stoichiometry of the gp41-gp59 complex. J Biol Chem 277:20555–62.
- Trotman LC et al. (2001) Import of adenovirus DNA involves the nuclear pore complex receptor can/nup214 and histone h1. Nat Cell Biol 3:1092–100.
- Horney MJ et al. (2001) Synthesis and characterization of insulin-like growth factor (igf)-1 photoprobes selective for the igf-binding proteins (igfbps). Photoaffinity labeling of the igf-binding domain on igfbp-2. J Biol Chem 276:2880–9.
- Daum JR et al. (2000) The spindle checkpoint of saccharomyces cerevisiae responds to separable microtubule-dependent events. Curr Biol 10:1375–8.
- Kleene R et al. (2000) Sh3 binding sites of zg29p mediate an interaction with amylase and are involved in condensation-sorting in the exocrine rat pancreas. Biochemistry 39:9893–900.
- Minami Y et al. (2000) A critical role for the proteasome activator pa28 in the hsp90-dependent protein refolding. J Biol Chem 275:9055–61.
- Sharma KK et al. (2000) Synthesis and characterization of a peptide identified as a functional element in aA-crystallin. J Biol Chem 275:3767–71.
- Ilver D et al. (1998) Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–7.
- Jacobson KA et al. (1995) Molecular probes for muscarinic receptors: Functionalized congeners of selective muscarinic antagonists. Life Sci 56:823–30.
For Research Use Only. Not for use in diagnostic procedures.
