Search
Ion 16S™ Metagenomics Solution

Ion 16S Metagenomics Kit and Ion Reporter metagenomics workflow solution
Culture-free, rapid identification of polybacterial research samples using Ion Torrent sequencing systems.
Ion semiconductor sequencing accelerates and simplifies metagenomics research through whole-genome or targeted sequencing of the bacterial 16S rRNA gene (16S sequencing). By eliminating the need to clone or culture samples prior to sequencing, one of the main biases in sampling is removed. Direct sequencing of samples on the Ion GeneStudio S5™ System or the Ion PGM™ System with Ion Hi-Q™ chemistry can be performed quickly, and with more amplicons, to enable more accurate discrimination between organisms down to genus and species levels.
The Ion 16S™ Metagenomics Kit is designed for rapid, comprehensive, and broad-range research analyses of mixed microbial populations. The kit uses two primer pools to amplify seven hypervariable regions (V2, V3, V4, V6, V7, V8, and V9) of bacterial 16S rRNA. The combination of the two primer pools enables broad-range, sequence-based identification of bacteria from complex mixed populations.
The amplified fragments can then be sequenced on the Ion GeneStudio S5 System or the Ion PGM System and analyzed using the metagenomics workflow in the Ion Reporter™ Software.
Video: The diversity of microbes
After decades of analyses and hundreds of studies, we’ve learned that the diversity of microbes is even greater than imagined—and the diversity of these species and their interactions may be linked to conditions such as IBD, obesity, and chronic wounds. Hear how Dr. George Watts and Dr. Charles Li are using Ion 16S sequencing in their clinical and environmental research applications.
Watch video ›
Benefits of the Ion 16S Metagenomics Solution include:
- Use directly on sample without culture, or on mixed or pure cultures
- Primer sets for multiple V regions allow accurate detection and identification of a broad range of bacteria down to genus or species level, utilizing Ion Reporter Software with the MicroSEQ™ ID 16S rRNA reference database and Greengenes database
- Flexibility—primer sets provided as two pools allow for separate analyses using individual pools
- Primers can also be barcoded
Ion 16S Metagenomics Kit and Ion Reporter metagenomics workflow flyer
Ion Reporter Metagenomics 16S algorithms white paper
Assessment of microbial population diversity in polymicrobial research samples by 16S single or multi-V region sequencing
Rapid 16S next-generation sequencing for bacterial identification in polymicrobial samples
ASM 2015 posters
Ion Torrent™ sequencing is cited in more than 900 peer-reviewed publications on microbial sequencing for bacterial and viral typing research, metagenomics, and discovery of unknown microorganisms.
View Ion Torrent publications >
Rapid 16S next-generation sequencing for bacterial identification in polymicrobial samples ›
Ion Torrent sequencing is cited in more than 900 peer-reviewed publications on microbial sequencing for bacterial and viral typing research, metagenomics, and discovery of unknown microorganisms
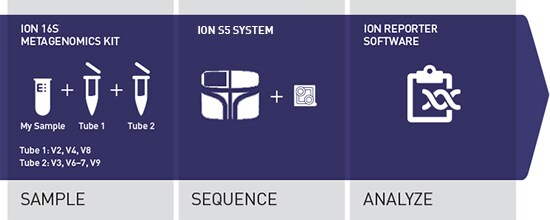
Integrated, cost-effective, scalable 16S rRNA sequencing workflow for the Ion GeneStudio S5 System, with a rapid workflow from library to analyzed results. Following library construction and template preparation, 400 bp sequencing runs are completed in just 4 hours for the Ion 520™ and Ion 530™ Chips. BAM files produced with Torrent Suite™ Software are automatically analyzed, annotated, and taxonomically assigned in the Ion Reporter Software 16S metagenomics workflow.

Ion GeneStudio S5 System
Adopting NGS in your lab is now simpler than ever before. The Ion GeneStudio S5 next-generation sequencing systems enable the simplest targeted sequencing workflow for your lab with industry-leading speed and affordability. Focus your research efforts on applications that have the potential to be impactful. The Ion GeneStudio S5 System has a modular build, so your lab has the flexibility to do multiple targeted sequencing applications on a single system.
Ion Reporter Software Metagenomics Workflow
Ion Reporter Software enables the identification, at the genus or species level, of microbes present in complex polybacterial samples, and uses both the premium curated MicroSEQ™ ID 16S rRNA reference database and the curated Greengenes database. The Ion Reporter Software Metagenomics Workflow also provides primer information, classification information, percent ID, and mapping information. With easy-to-use interactive Krona chart displays, you can interpret population diversity for your research at any taxonomic level.

Automated analysis, annotation, and taxonomical assignment implemented in Ion Reporter Software. Consensus view at the genus level of the balanced mock bacterial sample using more stringent analysis parameters. Multiple taxonomical assignments (slash calls) at the genus level are circled in red.
For Research Use Only. Not for use in diagnostic procedures.
