Search
High Throughput Delivery of siRNAs: Examples
In the article, High Throughput siRNA Delivery In Vitro: From Cell Lines to Primary Cells, we examined the efficiency and reproducibility of two techniques for high throughput delivery of siRNAs: reverse transfection and 96 well electroporation. We found that reverse transfection is compatible with most immortalized adherent cell types, whereas electroporation is superior for delivering siRNAs to most primary cells and cells grown in suspension. Once optimized, these methods support high cell viability while effectively delivering siRNAs. Finally, while maximum transfection efficiency and cell viability are cell type specific, siRNA delivery by both methods is highly reproducible. The adaptation of reverse transfection and electroporation for medium to high throughput applications makes it possible to deliver sets, or libraries, of siRNAs to virtually any cell type, enabling reverse genetics analyses of tens to thousands of genes per experiment.
Here we use reverse transfection of 174 different siRNAs in a functional genomic screen to identify kinase genes involved in cell proliferation. Data from this article and the previous one validating these high throughput delivery methods have been published in RNA (2005) 11:985–993 [1]. This analysis provides an example of the types of RNAi screening experiments that can be designed and demonstrates that these screens are feasible in any laboratory with a multichannel pipettor.
Here we use reverse transfection of 174 different siRNAs in a functional genomic screen to identify kinase genes involved in cell proliferation. Data from this article and the previous one validating these high throughput delivery methods have been published in RNA (2005) 11:985–993 [1]. This analysis provides an example of the types of RNAi screening experiments that can be designed and demonstrates that these screens are feasible in any laboratory with a multichannel pipettor.
Identifying Kinases Involved in Cell Proliferation
The goal of this siRNA library screen was to identify kinase genes that play a role in cell proliferation. To measure the effects of siRNA induced knockdown on cell proliferation, we monitored the number of live cells using an alamarBlue® (Biosource) microplate assay. An indicator for metabolic cell function, alamarBlue is processed by live cells into a digestion product that fluoresces and can be monitored using a fluorescence plate reader.
Reverse transfection was used to deliver 174 siRNAs targeting 58 different human kinases (3 distinct siRNAs per target gene) plus positive and negative control siRNAs into HeLa cells in 96 well plates. Transfection complexes were prepared in Opti-MEM® serum-free medium (Invitrogen) by mixing 0.3 µl siPORT™ NeoFX™ Transfection Agent and 1 pmol siRNA (Silencer® CMGC Kinase siRNA Library Subset). HeLa cells were then plated in 96 well plates with simultaneous addition of the transfection complexes. Transfections were performed in triplicate. Using a multichannel pipettor, reverse transfection of the 531 samples (174 kinase gene siRNAs + 3 control siRNAs done in triplicate = 531) took under two hours, including cell preparation. 72 hours after transfection, cell proliferation was measured by alamarBlue assay. Individual data points were compared to the results from the negative control non-silencing siRNA (Figure 1).
A subset of samples were analyzed by real-time RT-PCR to quantify target mRNA knockdown. For these samples, total RNA was isolated using the MagMAX-96 Total RNA Isolation Kit 24 hours after transfection. The purified, DNase-treated RNA was reverse transcribed with random decamers using the RETROscript® Kit. Gene expression levels were then determined by real-time PCR on the ABI Prism® 7900 SDS (Applied Biosystems). 18S rRNA was amplified as an endogenous control to adjust for well-to-well variation in the amount of starting template. The corrected values were normalized to average values obtained from samples individually transfected with three different non-silencing siRNAs ( Silencer Negative Control #1, #2, #3 siRNA).
The screen identified several kinases whose down-regulation appeared to inhibit or enhance cell proliferation. For several of the kinase genes, cell number per well differed significantly for each of the three siRNAs directed to the same target. These siRNAs reduced target mRNA expression to similar levels (data not shown). Given that siRNAs can target unintended genes [2], we suspect that in cases where only a single siRNA caused an effect, the siRNA targeted a distinct gene essential for cell proliferation in addition to the intended kinase. This result illustrates the advantage of using three different siRNAs per target in the screen. Indeed, in most cases two or three siRNAs altered cell numbers to a similar degree, and we were able to conclude that knocking down these kinases directly affected cell number.
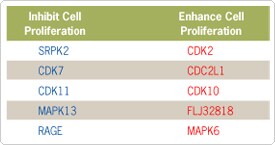
The screen identified 5 genes for which two or three siRNAs to that target resulted in lower than normal cell numbers (Figure 2). Knockdown of these genes could be due to a cell proliferation defect or other cellular mechanism. The screen also identified 5 genes that when knocked down by at least two different siRNAs resulted in higher than normal cell numbers. Several of the identified genes, such as CDK7, CDK11, CDK2, CDC2L1, and RAGE, have already been implicated in cell cycle regulation [3–7].
Reverse transfection was used to deliver 174 siRNAs targeting 58 different human kinases (3 distinct siRNAs per target gene) plus positive and negative control siRNAs into HeLa cells in 96 well plates. Transfection complexes were prepared in Opti-MEM® serum-free medium (Invitrogen) by mixing 0.3 µl siPORT™ NeoFX™ Transfection Agent and 1 pmol siRNA (Silencer® CMGC Kinase siRNA Library Subset). HeLa cells were then plated in 96 well plates with simultaneous addition of the transfection complexes. Transfections were performed in triplicate. Using a multichannel pipettor, reverse transfection of the 531 samples (174 kinase gene siRNAs + 3 control siRNAs done in triplicate = 531) took under two hours, including cell preparation. 72 hours after transfection, cell proliferation was measured by alamarBlue assay. Individual data points were compared to the results from the negative control non-silencing siRNA (Figure 1).

Figure 1. Screen for siRNAs Targeting Kinases That Differentially Affect Cell Proliferation. HeLa cells (5000 cells/well) were plated in 96 well plates simultaneously with addition of transfection complexes, prepared in Opti-MEM® serum-free medium (Invitrogen), by mixing 0.3 µl siPORT™ NeoFX™ Transfection Agent and 1 pmol siRNA (Silencer® CMGC Kinase siRNA Library Subset). 3 different siRNAs targeting each kinase were independently transfected in triplicate. Data was normalized with a negative control—the average of 3 non-silencing siRNAs (Silencer Negative Control #1, #2, #3 siRNA). Cells were incubated for 72 hours, and cell proliferation was measured using an alamarBlue® cell proliferation assay (Biosource) according to manufacturer recommended protocol. The average of the triplicate transfection for each of the three different siRNAs is plotted.
A subset of samples were analyzed by real-time RT-PCR to quantify target mRNA knockdown. For these samples, total RNA was isolated using the MagMAX-96 Total RNA Isolation Kit 24 hours after transfection. The purified, DNase-treated RNA was reverse transcribed with random decamers using the RETROscript® Kit. Gene expression levels were then determined by real-time PCR on the ABI Prism® 7900 SDS (Applied Biosystems). 18S rRNA was amplified as an endogenous control to adjust for well-to-well variation in the amount of starting template. The corrected values were normalized to average values obtained from samples individually transfected with three different non-silencing siRNAs ( Silencer Negative Control #1, #2, #3 siRNA).
The screen identified several kinases whose down-regulation appeared to inhibit or enhance cell proliferation. For several of the kinase genes, cell number per well differed significantly for each of the three siRNAs directed to the same target. These siRNAs reduced target mRNA expression to similar levels (data not shown). Given that siRNAs can target unintended genes [2], we suspect that in cases where only a single siRNA caused an effect, the siRNA targeted a distinct gene essential for cell proliferation in addition to the intended kinase. This result illustrates the advantage of using three different siRNAs per target in the screen. Indeed, in most cases two or three siRNAs altered cell numbers to a similar degree, and we were able to conclude that knocking down these kinases directly affected cell number.
The screen identified 5 genes for which two or three siRNAs to that target resulted in lower than normal cell numbers (Figure 2). Knockdown of these genes could be due to a cell proliferation defect or other cellular mechanism. The screen also identified 5 genes that when knocked down by at least two different siRNAs resulted in higher than normal cell numbers. Several of the identified genes, such as CDK7, CDK11, CDK2, CDC2L1, and RAGE, have already been implicated in cell cycle regulation [3–7].
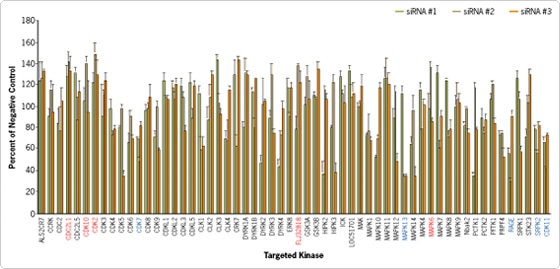
Figure 2. Kinase Genes Identified by the Screen. siRNAs to these 10 Kinase targets significantly decreased or increased cell numbers compared to negative control siRNA transfected cells.
Discussion
Double-stranded RNA library screening has enabled the classification of genes involved in apoptosis, phagocytosis, cell proliferation, the cell cycle, p53-mediated senescence, cell morphogenesis, cytokinesis, and signaling [8–14]. The availability of libraries of siRNAs has made it possible to evaluate the effects of sequentially reducing the expression of individual genes on cellular processes, providing a powerful reverse genetics tool for use in human and rodent cells. Key to the effective use of siRNA libraries in screening studies are procedures that enable hundreds or thousands of active siRNAs to be delivered to cells with high efficiency while minimizing toxicity in the cells associated with the delivery method.
The NCBI compendium currently lists over 5500 human genes with unknown function. Thus, siRNA screening experiments provide an obvious opportunity for gene function discovery. There are dozens of enzymatic and biological assays commercially available to aid in phenotypic screening. Ambion has worked to develop reagents and techniques that simplify the high throughput siRNA delivery process, and to make it accessible to even small laboratories without robotic capabilities. In addition to providing several highly effective siRNAs to each human, mouse, and rat gene (over 200,000 siRNAs), Ambion offers a wide selection of siRNA libraries to human gene targets, grouped by functional class
The NCBI compendium currently lists over 5500 human genes with unknown function. Thus, siRNA screening experiments provide an obvious opportunity for gene function discovery. There are dozens of enzymatic and biological assays commercially available to aid in phenotypic screening. Ambion has worked to develop reagents and techniques that simplify the high throughput siRNA delivery process, and to make it accessible to even small laboratories without robotic capabilities. In addition to providing several highly effective siRNAs to each human, mouse, and rat gene (over 200,000 siRNAs), Ambion offers a wide selection of siRNA libraries to human gene targets, grouped by functional class
References
- Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D. (2005) High-throughput RNAi screening in vitro: From Cell Lines to primary cells. RNA11:985–93.
- Jackson AL, Linsley PS (2004) Noise amidst the silence: off-target effects of siRNAs? Trends Genet 20:521–4.
- Pagano M, Draetta G, Jansen-Durr P (1992) Association of cdk2 kinase with the transcription factor E2F during S phase. Science 255:1144–7.
- Liang YC, Tsai SH, Chen L, Lin-Shiau SY, Lin JK (2003) Resveratrol-induced G2 arrest through the inhibition of CDK7 and p34CDC2 kinases in colon carcinoma HT29 cells. Biochem Pharmacol 65:1053–60.
- Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW, Nelson MA (2003) The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem 278:5062–71.
- Feng Y, Goulet AC, Nelson MA (2004) Identification and characterization of the human Cdc2l2 gene promoter. Gene 330:75–84.
- Brizzi MF, Dentelli P, Rosso A, Calvi C, Gambino R, Cassader M, Salvidio G, Deferrari G, Camussi G, Pegoraro L, Pagano G, Cavallo-Perin P (2004) RAGE- and TGF-beta receptor-mediated signals converge on STAT5 and p21waf to control cell-cycle progression of mesangial cells: a possible role in the development and progression of diabetic nephropathy. FASEB J 18:1249–51.
- Sachse C, Echeverri CJ (2004) Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene 23:8384–91.
- Somma MP, Fasulo B, Cenci G, Cundari E, Gatti M (2002) Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol Biol Cell 13:2448–60.
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N (2003) A functional genomic analysis of cell morphology using RNA interference. J Biol 2:27.
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA (2003) Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299:2039–45.
- Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL (2004) Differential Regulation of the TRAIL Death Receptors DR4 and DR5 by the Signal Recognition Particle. Mol Biol Cell 15(11):5064–74.
- Xin H, Bernal A, Amato FA, Pinhasov A, Kauffman J, Brenneman DE, Derian CK, Andrade-Gordon P, Plata-Salaman CR, Ilyin SE. (2004) High-throughput siRNA-based functional target validation. J Biomol Screen 9(4):286–93.
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature436:78–86.