Search Thermo Fisher Scientific
Metabolic Labeling Strategies
Page contents
View and select products
Radioactive metabolic labeling
Radioactive analogs are the most commonly used compounds for metabolic labeling of cells and organisms. Since radiolabeled isotopes can be substituted in biomolecule monomers without any changes to the chemical structure, they are readily incorporated in vivo. Radiolabeled macromolecules are also easily detected by sensitive radiometric techniques such as liquid scintillation counting or positron emission tomography (PET) scanning. Examples of radioactive tracers and applications include 3H thymidine uptake for cell proliferation assays, 35S methionine labeling for protein synthesis determination, 32P orthophosphate labeling for in vivo kinase assays, and 14C-labeled D-glucose update for determination of cellular metabolism rates. Although radioactive isotopes are easily detected and relatively inexpensive, there are some disadvantages including safety hazards, generation of radioactive waste, toxicity to organisms, and radioactive decay leading to loss of signal over time.
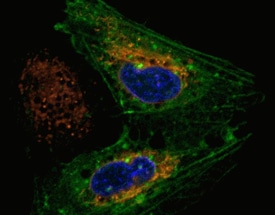
This representative experiment shows how azide-reactive Thermo Scientific DyLight dyes may be used to label cultured cells.
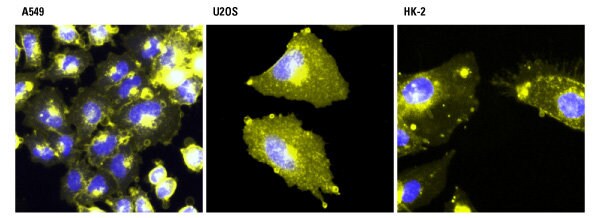
Detection of metabolic labeling with azido-sugars in different cell types using azide-reactive DyLight dyes. A549, U2OS and HK-2 cells were incubated with 40 µM azido-acetylmannosamine in cell culture media for 72 hours and then incubated with 100 µM of Thermo Scientific DyLight 550-Phosphine (yellow). The cells were washed, fixed with 4% paraformaldehyde and counterstained with Thermo Scientific Pierce Hoechst 33342 (blue).
Bioconjugate Techniques, 3rd Edition
Bioconjugate Techniques, 3rd Edition (2013) by Greg T. Hermanson is a major update to a book that is widely recognized as the definitive reference guide in the field of bioconjugation.
Bioconjugate Techniques is a complete textbook and protocols-manual for life scientists wishing to learn and master biomolecular crosslinking, labeling and immobilization techniques that form the basis of many laboratory applications. The book is also an exhaustive and robust reference for researchers looking to develop novel conjugation strategies for entirely new applications. It also contains an extensive introduction to the field of bioconjugation, which covers all the major applications of the technology used in diverse scientific disciplines, as well as tips for designing the optimal bioconjugate for any purpose.
Stable isotope labeling using SILAC
In contrast to radioactive metabolic labeling, stable isotope labeling uses isotopic analogs of biomolecule monomers which are not radioactive. Like radioisotopes, stable isotopes can be incorporated into biomolecule monomers without any changes to the chemical structure. Common stable isotopes used for metabolic labeling include 2H, 15N, 13C and 18O. Since they are not radioactive, these heavy stable isotopes are detected by their increased mass compared to natural light isotopes using mass spectrometry (MS).
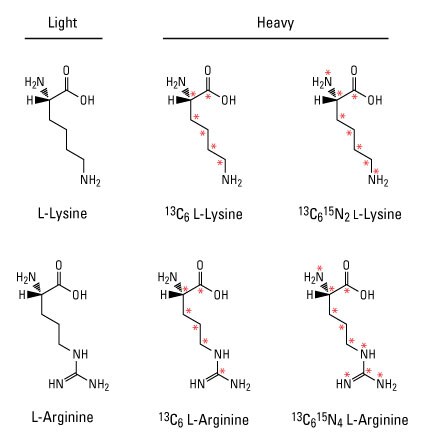
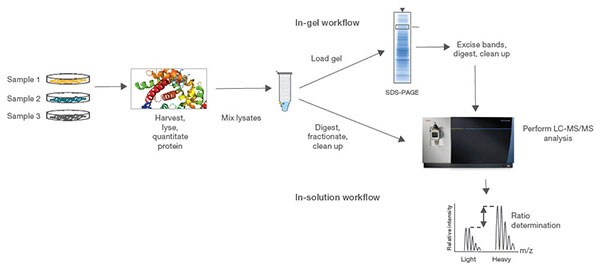
Stable isotope labeling by amino acids in cell culture (SILAC) is the most common approach for metabolic labeling using stable isotopes. In this method, cells are cultured in special growth medium that contains either light or heavy amino acids such as 13C6-lysine and 13C6-arginine. After labeling, light and heavy samples are combined to measure differences in relative protein expression or post-translational modifications by MS. This powerful quantitative proteomic technique is particularly useful to detect relatively small changes in protein levels and can be multiplexed to measure up to three different experimental conditions in a single MS analysis.
Learn more
- Quantitative Proteomics
- Handbook: Thermo Scientific Pierce Reagents for Quantitative Proteomics
- Reference manual: Quantitative Protein Expression Technologies
- Protein Mass Spectrometry Information
- Protein Assays and Analysis Support Center
Select products
Photoreactive amino acids
Photoreactive amino acids are chemical analogs of amino acids that enable metabolic labeling and in vivo crosslinking. These amino acids contain a small diazirine functional group which is stable under normal lab environments but can be photoactivated with long-wave UV light (330 to 370 nm). Upon UV activation, the diazirine group is liberated to generate reactive carbene intermediates, which covalently bond to any amino acid side chain or peptide backbone in close proximity.
Photo-L-Leucine and Photo-L-Methionine are two examples of photoreactive amino acids designed for metabolic labeling of proteins. An advantage of using these amino acids for studying protein–protein interactions is the ability to crosslink proteins at their protein interaction interfaces. However, since these amino acids are not identical in structure to their natural counterparts, photoreactive amino acids typically have lower incorporation rates than isotopic analogs and are not well tolerated by cells or complex organisms for long growth periods.
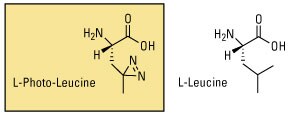
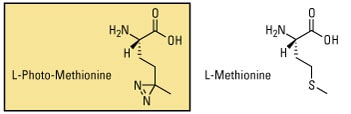
Learn more
- Photoreactive Crosslinker Chemistry
- Tech Tip #11: Light sources and conditions for photoactivation of aryl azide crosslinking and labeling reagents
Select products
Bioorthogonal labeling reagents
Bioorthogonal labeling reagents are metabolic labeling compounds whose functional groups can be chemically reacted without interfering with biological processes. Bioorthogonal functional groups that have been used as the basis for metabolic labeling compounds include azides, alkynes, aldehydes and ketones. Unlike radioactive or stable isotope analogs, bioorthogonal analogs are not detected directly but rather reacted with another detection or affinity compound through a chemoselective ligation reaction.
One chemoselective ligation reaction is a copper-catalyzed Huisgen cycloaddition between azides and alkynes, commonly referred to as Click chemistry. Either azides or alkynes can be used as the functional group for metabolic labeling due to their small size and compatibility with biological systems. However, traditional Click chemistry reactions use copper as a catalyst, which is toxic to living organisms. Therefore, copper-free Click probes using larger cycloalkynes have been developed as an alternative method for ligation.
The Staudinger ligation is another chemoselective chemistry which involves the reaction of azido bioorthogonal probes with phosphine compounds. Similar to Click chemistry, the ligation reaction is highly specific and can be performed in aqueous environments at physiological pH. However, unlike traditional Click chemical reactions, Staudinger ligations do not require copper to be reactive. Although this increases the biocompatibility of Staudinger ligations, these reactions also tend to be slower than Click reactions due to the absence of a catalyst.
Click chemistry is the detection method of choice for samples that would be compromised by direct labeling or antibody-based secondary detection techniques. The click label is small enough to penetrate complex samples easily, and the selectivity and stability of the Click reaction provides high sensitivity and low background signal. This gentle sample treatment together with the biocompatible Invitrogen Click-iT Plus reaction means that detection can be multiplexed with expressed proteins such as GFP, protein labels such as R-PE, and a wide range of organic fluorophores.
Watch this video to learn more about Click chemistry
Learn more
- Chemoselective Ligation Reaction Chemistry
- Click Chemistry—Section 3.1
- Dual Pulse Labeling of Cell Proliferation
- Technical Reference: New Click-iT DIBO Tools Preserve Cell Viability and Protein Function With Copper-Free Protein Labeling and Detection
Bioorthogonal probes
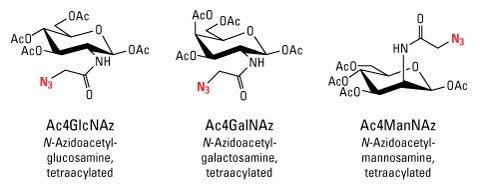
Numerous bioorthogonal probes have been developed for metabolic labeling, including azidohomoanaline for labeling newly synthesized proteins, 5-ethynyl-2'-deoxyuridine (EdU) for labeling nucleic acids, azido palmitic acid for labeling inter-membrane proteins, and azido sugars for labeling glycoproteins. Azido sugar reagents were one of the first bioorthogonal probes used for metabolic labeling. After incorporation of the azido sugar into N-linked glycans, they can be chemoselectively targeted with alkyne- or phosphine-derivatized compounds for detection or affinity purification.
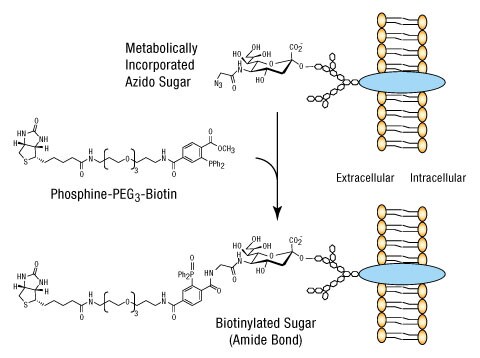
Due to the wide variety of detection and affinity compounds available for ligation ranging from biotins to fluorescent dyes to mass tags, bioorthogonal labeling is one of the most versatile methods for metabolically labeling biomolecules. The illustration provides an example of how biotinylated sugar moieties may be used for in vivo labeling.
Learn more
- Protein Glycosylation
- Application Note: Click-iT Metabolic Labeling Reagents for Proteins
- Labeling Chemistry Support
- DyLight Fluors Technology and Product Guide
Select products
- GlcNAz (N-azidoacetylglucosamine tetraacylated)
- ManNAz (N-azidoacetylmannosamine tetraacylated)
- GalNAz (N-azidoacetylgalactosamine tetraacylated)
- EdU (5-ethynyl-2’-deoxyuridine)
- EU (5-Ethynyl Uridine)
- Click-iT Palmitic Acid, Azide (15-Azidopentadecanoic Acid)
- Click-iT Labeling and Detection Tools
- EZ-Link Phosphine-PEG3-Biotin
- DyLight 650-Phosphine
- EZ-Link Phosphine-PEG4-Desthiobiotin
Site-specific metabolic labeling
Metabolic labeling of biomolecules distributes the label throughout the molecule based on the biomolecule composition. For example, culturing cells with 13C6-lysine results in the incorporation (labeling) of all lysine-containing proteins synthesized during the experiment. Because most metabolic labeling strategies use chemical analogs which are structurally identical or very similar to natural monomers, it is usually not possible to control the specific site of incorporation or the number of labels added. However, site-specific metabolic labeling is possible when metabolic probes are designed and used in combination with genetically altered organisms or cell-free in vitro translation systems.
One method for site-specific incorporation takes advantage of redundant stop codons present in the genetic code, which can be used to incorporate amino acid analogs using specifically engineered tRNAs. In addition to allowing for site-specific incorporation, this method allows for labeling with chemical analogs such as biotinylated or fluorescent dye-labeled amino acids which are not tolerated by endogenous synthesis and modification machinery. This method also limits the incorporation of the metabolic label to the expressed, genetically modified proteins.
Fluorescent probes used in live cells
Fluorescent molecules, also called fluorophores or simply fluors, respond directly and distinctly to light and produce a detectable signal. Unlike enzymes or biotin, fluorescent labels do not require additional reagents for detection. This feature makes fluorophores extremely versatile and the new standard in detecting protein location and activation, identifying protein complex formation and conformational changes, and monitoring biological processes in vivo.
The vast selection of fluorophores today provides greater flexibility, variation and fluorophore performance for research applications than ever before. Fluorophores can be divided into three general groups, and each group of probes has distinct characteristics. These groups are as follows:
- Organic dyes—FITC, TRITC, DyLight fluors
- Biological fluorophores—Green fluorescent protein (GFP), R-Phycoerythrin
- Quantum dots
Live Cell Imaging eBook
Published by Wiley, this free eBook provides essential knowledge and provides a short, practical guide of the background and basics of live cell imaging. Content includes:
- Basic history of live cell imaging
- Case studies of live cell imaging applications
- Common problems and solutions with live cell imaging

Select products
- Endoplasmic Reticulum (ER) Structure
- Golgi Complex Structure Selection Guides
- CellEvent Caspase-3/7 Green Detection Reagent
- CellLight Early Endosomes-GFP, BacMam 2.0
- Qtracker 655 Cell Labeling Kit
- CellLight Peroxisome-GFP, BacMam 2.0
- CellLight Histone 2B-GFP, BacMam 2.0
- MitoTracker Red CM-H2Xros - Special Packaging
- CellLight Lysosomes-RFP, BacMam 2.0
- LIVE/DEAD BacLight Bacterial Viability Kit, for microscopy
- Live Cell Imaging of Cell Cycle and Division
Recommended reading
- Ong SE et al. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1(5):376–86.
- Suchanek M et al. (2005) Photo-leucine and photo-methionine allow identification of protein-protein interactions. Nat Methods 2(4):261–267.
- Prescher JA, Bertozzi CR (2005) Chemistry in living systems. Nat Chem Biol 1(1):13–21.
- Kolb HC et al. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl 40(11):2004–2021.
- Agard NJ et al. (2006) A comparative study of bioorthogonal reactions with azides. ACS Chem Biol 1(10):644–648.
- Saxon E, Bertozzi C (2000) Cell surface engineering by a modified Staudinger reaction. Science 287(5460):2007–10.
- Wang L et al. (2001) Expanding the genetic code of Escherichia coli. Science 292 (5516):498–500.
- Crivat G, Taraska JW (2012) Imaging proteins inside cells with fluorescent tags. Trends Biotechnol 30:8–16.
- Horan PK et al. (1990) Fluorescent cell labeling for in vivo and in vitro cell tracking. Methods Cell Biol 33:469–490.
For Research Use Only. Not for use in diagnostic procedures.