Search
Phosphorylation
Reversible protein phosphorylation, principally on serine, threonine or tyrosine residues, is one of the most important and well-studied post-translational modifications. Phosphorylation plays critical roles in the regulation of many cellular processes including cell cycle, growth, apoptosis and signal transduction pathways.
Phosphorylation is the most common mechanism of regulating protein function and transmitting signals throughout the cell. While phosphorylation has been observed in bacterial proteins, it is considerably more pervasive in eukaryotic cells. It is estimated that one-third of the proteins in the human proteome are substrates for phosphorylation at some point. Indeed, phosphoproteomics has been established as a branch of proteomics that focuses solely on the identification and characterization of phosphorylated proteins.
Mechanism of phosphorylation
While phosphorylation is a prevalent post-translational modification (PTM) for regulating protein function, it only occurs at the side chains of three amino acids, serine, threonine and tyrosine, in eukaryotic cells. These amino acids have a nucleophilic (–OH) group that attacks the terminal phosphate group (γ-PO32-) on the universal phosphoryl donor adenosine triphosphate (ATP), resulting in the transfer of the phosphate group to the amino acid side chain. This transfer is facilitated by magnesium (Mg2+), which chelates the γ- and β-phosphate groups to lower the threshold for phosphoryl transfer to the nucleophilic (–OH) group. This reaction is unidirectional because of the large amount of free energy that is released when the phosphate–phosphate bond in ATP is broken to form adenosine diphosphate (ADP).
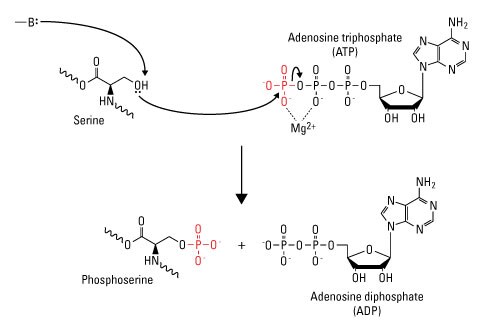
Diagram of serine phosphorylation. Enzyme-catalyzed proton transfer from the (–OH) group on serine stimulates the nucleophilic attack of the γ-phosphate group on ATP, resulting in transfer of the phosphate group to serine to form phosphoserine and ADP. (—B:) indicates the enzyme base that initiates proton transfer.
For a large subset of proteins, phosphorylation is tightly associated with protein activity and is a key point of protein function regulation. Phosphorylation regulates protein function and cell signaling by causing conformational changes in the phosphorylated protein. These changes can affect the protein in two ways. First, conformational changes regulate the catalytic activity of the protein. Thus, a protein can be either activated or inactivated by phosphorylation. Second, phosphorylated proteins recruit neighboring proteins that have structurally conserved domains that recognize and bind to phosphomotifs. These domains show specificity for distinct amino acids. For example, Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains show specificity for phosphotyrosine (pY), although distinctions in these two structures give each domain specificity for distinct phosphotyrosine motifs. Phosphoserine (pS) recognition domains include MH2 and the WW domain, while phosphothreonine (pT) is recognized by forkhead-associated (FHA) domains. The ability of phosphoproteins to recruit other proteins is critical for signal transduction, in which downstream effector proteins are recruited to phosphorylated signaling proteins.
Protein phosphorylation is a reversible PTM that is mediated by kinases and phosphatases, which phosphorylate and dephosphorylate substrates, respectively. These two families of enzymes facilitate the dynamic nature of phosphorylated proteins in a cell. Indeed, the size of the phosphoproteome in a given cell is dependent upon the temporal and spatial balance of kinase and phosphatase concentrations in the cell and the catalytic efficiency of a particular phosphorylation site.
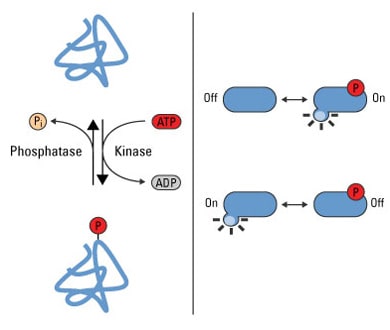
Phosphorylation is a reversible PTM that regulates protein function. Left panel: Protein kinases mediate phosphorylation at serine, threonine and tyrosine side chains, and phosphatases reverse protein phosphorylation by hydrolyzing the phosphate group. Right panel: Phosphorylation causes conformational changes in proteins that either activate (top) or inactivate (bottom) protein function.
This 118-page handbook provides comprehensive information about protein expression and will help you choose the right expression system and purification technologies for your specific application and needs. Get tips and tricks when starting an experiment, and find answers to everyday problems related to protein expression.

Learn more
- Overview of Protein-Protein Interaction Analysis
- Phosphoproteomics
- Overview of Post-translational Modifications (PTMs)
- Application Note: Phosphoprotein Enrichment from Cell and Tissue Samples
- Protein Expression Support Center
Kinases are enzymes that facilitate phosphate group transfer to substrates. Greater than 500 kinases have been predicted in the human proteome; this subset of proteins comprises the human kinome. Substrates for kinase activity are diverse and include lipids, carbohydrates, nucleotides and proteins.
ATP is the cosubstrate for almost all protein kinases, although guanosine triphosphate is used by a small number of kinases. ATP is the ideal structure for the transfer of α-, β- or γ-phosphate groups for nucleotidyl-, pyrophosphoryl- or phosphoryltransfer, respectively. While the substrate specificity of kinases varies, the ATP-binding site is generally conserved.
Protein kinases are categorized into subfamilies that show specificity for distinct catalytic domains and include tyrosine kinases or serine/threonine kinases. Approximately 80% of the mammalian kinome comprises serine/threonine kinases, and >90% of the phosphoproteome consists of pS and pT. Indeed, studies have shown that the relative abundance ratio of pS:pT:pY in a cell is 1800:200:1. Although pY is not as prevalent as pS and pT, global tyrosine phosphorylation is at the forefront of biomedical research because of its relation to human disease via the dysregulation of receptor tyrosine kinases (RTKs).
Protein kinase substrate specificity is based not only on the target amino acid but also on consensus sequences that flank it. These consensus sequences allow some kinases to phosphorylate single proteins and others to phosphorylate multiple substrates (>300). Additionally, kinases can phosphorylate single or multiple amino acids on an individual protein if the kinase-specific consensus sequences are available.
Kinases have regulatory subunits that function as activating or autoinhibitory domains and have various regulatory substrates. Phosphorylation of these subunits is a common approach to regulating kinase activity. Most protein kinases are dephosphorylated and inactive in the basal state and are activated by phosphorylation. A small number of kinases are constitutively active and are made intrinsically inefficient, or inactive, when phosphorylated. Some kinases, such as Src, require a combination of phosphorylation and dephosphorylation to become active, indicating the high regulation of this proto-oncogene. Scaffolding and adaptor proteins can also influence kinase activity by regulating the spatial relationship between kinases and upstream regulators and downstream substrates.
The activity of specific kinases can be measured by incubating immunoprecipitates with substrates for specific kinases and ATP. Commercial kits are available for this type of assay and are designed to yield colorimetric, radiometric or fluorometric detection. While this type of assay shows the activity of specific kinases, as with kinase enrichment, it does not provide information on the proteins that the kinases modify or the role of endogenous phosphatase activity.
The reversibility of protein phosphorylation makes this type of PTM ideal for signal transduction, which allows cells to rapidly respond to intracellular or extracellular stimuli. Signal transduction cascades are characterized by one or more proteins physically sensing cues, either through ligand binding, cleavage or some other response, that then relay the signal to second messengers and signaling enzymes. In the case of phosphorylation, these receptors activate downstream kinases, which then phosphorylate and activate their cognate downstream substrates, including additional kinases, until the specific response is achieved. Signal transduction cascades can be linear, in which kinase A activates kinase B, which activates kinase C and so forth. Signaling pathways have also been discovered that amplify the initial signal; kinase A activates multiple kinases, which in turn activate additional kinases. With this type of signaling, a single molecule, such as a growth factor, can activate global cellular programs such as proliferation.
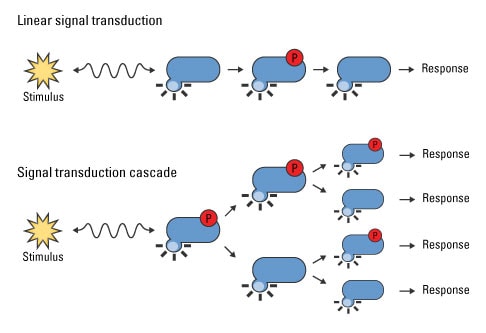
Signal transduction cascades amplify the signal output. External and internal stimuli induce a wide range of cellular responses through a series of second messengers and enzymes. Linear signal transduction pathways yield the sequential activation of a discrete number of downstream effectors, while other stimuli elicit signal cascades that amplify the initial stimulus for large-scale or global cellular responses.
The intensity and duration of phosphorylation-dependent signaling is regulated by three mechanisms:
- Removal of the activating ligand
- Kinase or substrate proteolysis
- Phosphatase-dependent dephosphorylation
The human proteome is estimated to contain approximately 150 protein phosphatases, which show specificity for pS/pT and pY residues. While dephosphorylation is the end goal of these two groups of phosphatases, they do it through separate mechanisms. Serine/threonine phosphatases mediate the direct hydrolysis of the phosphorus atom of the phosphate group using a bimetallic (Fe/Zn) center, while tyrosine phosphatases form a covalent thiophosphoryl intermediate that facilitates removal of the tyrosine residue.
Because of the influence that phosphorylation has on biological processes in general, a huge emphasis has been placed on understanding the biological role of protein phosphorylation in the context of human disease. Small-scale protein phosphorylation is commonly performed to study the activity of a small number of proteins, while phosphoproteomic analyses are increasingly used to understand the global dynamics of phosphorylation of entire protein families. Current approaches to study protein phosphorylation include immunodetection, phosphoprotein or phosphopeptide enrichment, kinase activity assays and mass spectrometry. Because of the detrimental effect that phosphatases can have on the detection of phosphorylated proteins, broad-spectrum phosphatase inhibitors are commonly added to cell lysates in many phosphodetection strategies.
Immunodetection
Phospho-specific antibodies raised against specific phospho-epitopes on target proteins are a core tool for studying site-specific protein phosphorylation. On a more global scale, antibodies have been developed to detect the phosphorylation of specific amino acids (pS, pT, pY). Phospho-specific antibodies can be used for traditional western blotting, immunoprecipitation (IP), immunohistochemistry (IHC), ELISA, flow cytometry and, more recently, immobilization onto solid support arrays. Although phospho-specific antibodies are popular amongst the research community, only a small fraction of them are highly specific for their targets and many have issues with low sensitivity.
Phosphorylation-specific ELISA kits. Phosphorylation-specific Invitrogen ELISA kits are high-quality ELISA kits, designed for researchers studying intracellular proteins involved in signaling pathways. These assay kits are designed to deliver accurate, sensitive, and fast protein quantitation of total and phosphorylated, modified, or cleavage site-specific proteins in a broad range of sample types.
In the following example, western blot analysis was used to detect the protein p38 MAPK, a serine/threonine kinase that plays an important role in signal transduction, contributing to the regulation of many cellular processes including cell differentiation and inflammation.
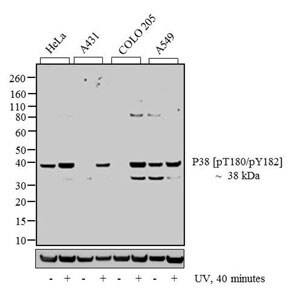
Detection of p38 MAP protein. Western blot analysis of p38 [pT180]/[pY182] was performed by loading 20 µg of HeLa (lane 1), HeLa exposed for 40 minutes with UV (lane 2), A431 (lane 3), A431 exposed for 40 minutes with UV (lane 4), COLO 205 (lane 5), COLO 205 exposed for 40 minutes with UV (lane 6), A549 (lane 7) and A549 exposed for 40 minutes with UV (lane 8) cell lysate using Invitrogen NuPAGE 4-12 % Bis-Tris Gel (Cat. No. NP0322BOX), XCell SureLock Electrophoresis System (EI0002), Novex Sharp Pre-Stained Protein Standard (LC5800), and iBlot Dry Blotting System (IB21001). Proteins were transferred to a nitrocellulose membrane and blocked with 5 % skim milk for 1 hour at room temperature. p38 [pT180]/[pY182] was detected at ~38 kDa using Invitrogen p38 [pT180]/[pY182] rabbit polyclonal antibody (44684G) at 1:1,000 in 5 % skim milk at 4°C overnight on a rocking platform. Invitrogen goat anti-rabbit IgG–HRP secondary antibody (G21234) at 1:5,000 dilution was used and chemiluminescent detection was performed using Invitrogen Novex ECL Chemiluminescent Substrate Reagent Kit (WP20005).
Enrichment
Key dynamic changes in protein phosphorylation occur on low abundance proteins, which require enrichment prior to proteomic analysis. These strategies include metal oxide affinity chromatography (MOAC) and immobilized metal affinity chromatography (IMAC), which use metal-ligand complexes to capture phosphate groups on pS, pT and pY. MOAC is most commonly performed with TiO2-chelated resins to form bidentate complexes with phosphates, while IMAC employs Fe-chelated support to form tri- or tetradentate complexes with phosphates. Commercial kits are also available that have proprietary phosphate-binding elements for phosphoprotein enrichment.
Another enrichment strategy is the elimination of the labile phosphate group (β-elimination) under strongly basic conditions. Phosphate groups are replaced with biotin moieties, and proteins/peptides are then enriched on avidin supports. A major drawback of this approach is the inability to β-eliminate the phosphate group of tyrosine residues.
Kinases (ATPases) and GTPases can also be enriched, although this approach does not provide information on the proteins that these enzymes modify. These enrichment strategies use nucleotide derivatives that bind to the active site of kinases or GTPases (depending on the derivative used) and mediate the covalent attachment of a modified biotin (desthiobiotin). Because desthiobiotin exhibits reversible binding to streptavidin, labeled kinases or GTPases can be enriched from samples.
Specific enzymes can be enriched using consensus sequences of downstream proteins as probes. This approach is popular with enriching GTPases, in which the protein-binding domains of the downstream effectors of specific GTPases are fused to GST for the selective enrichment of distinct GTPases.
The following table provides information about the performance of the Thermo Scientific Pierce Phosphoprotein Enrichment Kit designed for the efficient enrichment of phosphorylated proteins derived from mammalian cells and tissues.
| Kit | Phosphoprotein Yield (µg)† | Enrichment Time (Hours) |
| Thermo Scientific Pierce Phosphoprotein Enrichment Kit | 300 | 1.5 |
| Supplier Q Kit | 88 | 4.5 |
| Supplier I Kit | 52†† | 3.5 |
| Supplier C Kit | 160 | 3 |
| Supplier E Kit | Too dilute to determine | 5 |
| †From 2mg total protein; ††Based on maximum 1mg load per manufacturer’s protocol | ||
Learn more
- Protein Purification Using Affinity Chromatography
- Protein Purification
- Application Note: GTPase Enrichment Using a New Active-Site Probe
Quantitative mass spectrometry
Recent mass spectrometric strategies using stable isotope labeling by amino acids in cell culture (SILAC) or labeling of peptides in vitro with tandem mass tags have paved the way for the relative determination of changes in phosphorylation.
Absolute quantitation strategies are also available using isotopically "heavy" peptide standards. These approaches allow researchers to understand global phosphoproteomic changes in response to different stimuli or disease states. The diagram below illustrates the nature of complexity associated with protein sample preparation for MS. Phosphopeptide enrichment reduces sample complexity and is required prior to MS due to the low stoichiometry and poor ionization of phosphopeptides. Phosphoprotein enrichment is enhanced with the aid commercially available kits.
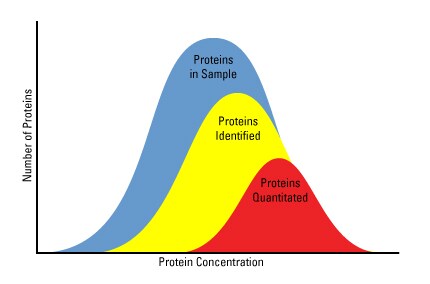
Protein availability for quantitative proteomic analysis is limited. Protein abundance and sample complexity are significant factors that affect the availability of proteins for mass spectrometric quantitation.
- Cohen P (2000) The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci 25:596–601.
- Yaffe MB (2002) Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 3:177–86.
- Bantscheff M et al. (2007) Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem 389:1017–31.
- Manning G et al. (2002) The protein kinase complement of the human genome. Science 298:1912–34.
- Walsh C (1979) Enzymatic reaction mechanisms. San Francisco (CA): W. H. Freeman. xv, p 978.
- Walsh C (2006) Posttranslational modification of proteins: Expanding nature's inventory. Englewood (CO): Roberts and Co. Publishers. xxi, p 490.
- Mann M et al. (2002) Analysis of protein phosphorylation using mass spectrometry: Deciphering the phosphoproteome. Trends Biotechnol 20:261–8.
- Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300:445–52.
- Johnson LN, Lewis RJ (2001) Structural basis for control by phosphorylation. Chem Rev 101:2209–42.
- Johnson GL, Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–2.
- Kennelly PJ (2001) Protein phosphatases--a phylogenetic perspective. Chem Rev 101:2291–312.
- Jackson MD, Denu JM (2001) Molecular reactions of protein phosphatases--insights from structure and chemistry. Chem Rev 101:2313–40.
- Ham BM (2012) Proteomics of biological systems: Protein phosphorylation using mass spectrometry techniques. Hoboken (NJ): John Wiley & Sons.
For Research Use Only. Not for use in diagnostic procedures.