Search Thermo Fisher Scientific

Platinum II Taq Hot-Start DNA PolymeraseRequest sample Resources |
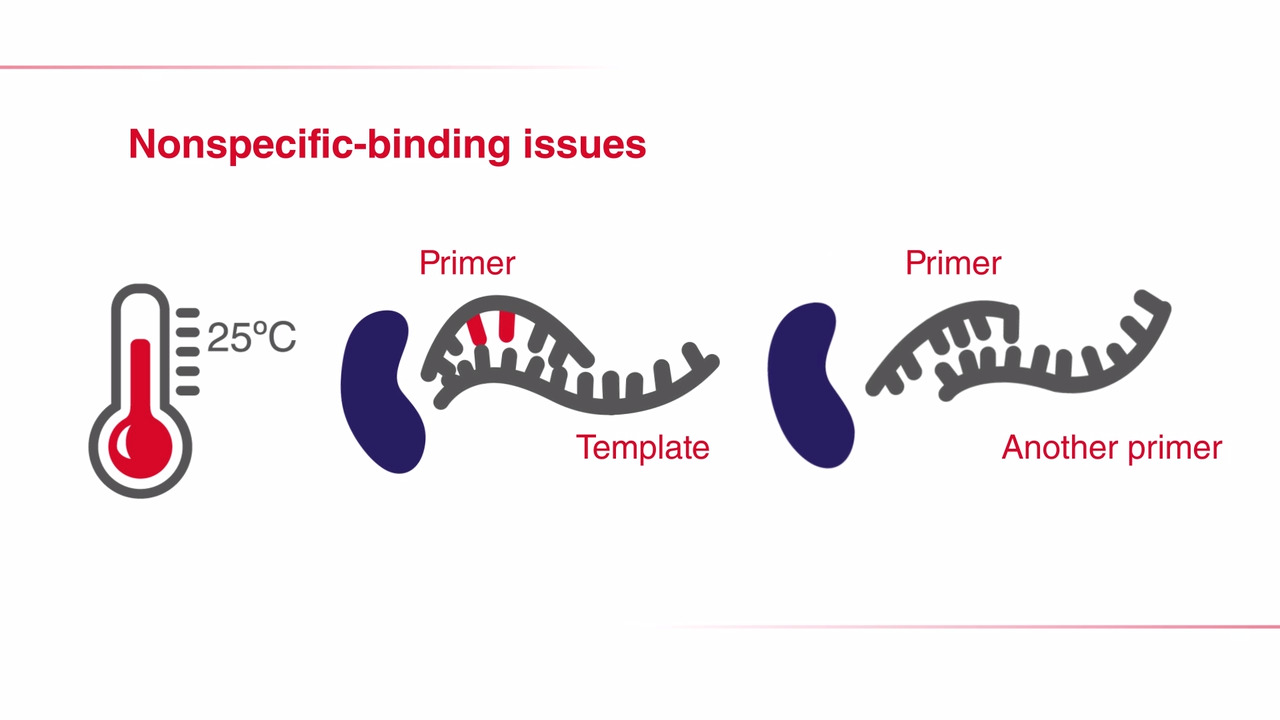
Hot-start PCR is a modified version of the traditional polymerase chain reaction (PCR) technique that provides higher specificity and sensitivity by minimizing non-specific amplification. In hot-start PCR, the polymerase stays inactive at lower temperatures due to an inhibitor or modification of the enzyme to prevent extension of primers at lower temperatures. Upon heating, the inhibitor is released and amplification begins.
Watch the video on common issues in traditional PCR amplification and how you can resolve them with hot-start PCR.
Highlights of Platinum II Taq Hot-Start DNA Polymerase
- Universal primer annealing at 60°C—No optimization of primer annealing temperatures and multiple PCR runs can be co-cycled together
- 4x faster DNA synthesis—Faster synthesis rates than traditional Taq polymerases
- High inhibitor tolerance—Utilizes an engineered Taq polymerase that is extremely robust
- Platinum hot-start technology—Enhances PCR specificity, sensitivity, and yields; allows for room temperature reaction setup
- Green buffer formats—Help reduce pipetting errors and enable direct gel loading
Advantages of Platinum II Taq Hot-Start DNA Polymerase
The innovative Platinum II PCR buffer enables universal primer annealing protocol by isostabilizing primer-template duplex structures. When amplifying with Platinum II Taq DNA Polymerase, a single 60°C annealing temperature can be used for any primer pair.

Watch the benefits of universal annealing feature.
Platinum II Taq Hot-Start DNA Polymerase is four times faster than conventional Taq DNA polymerase. Watch video.
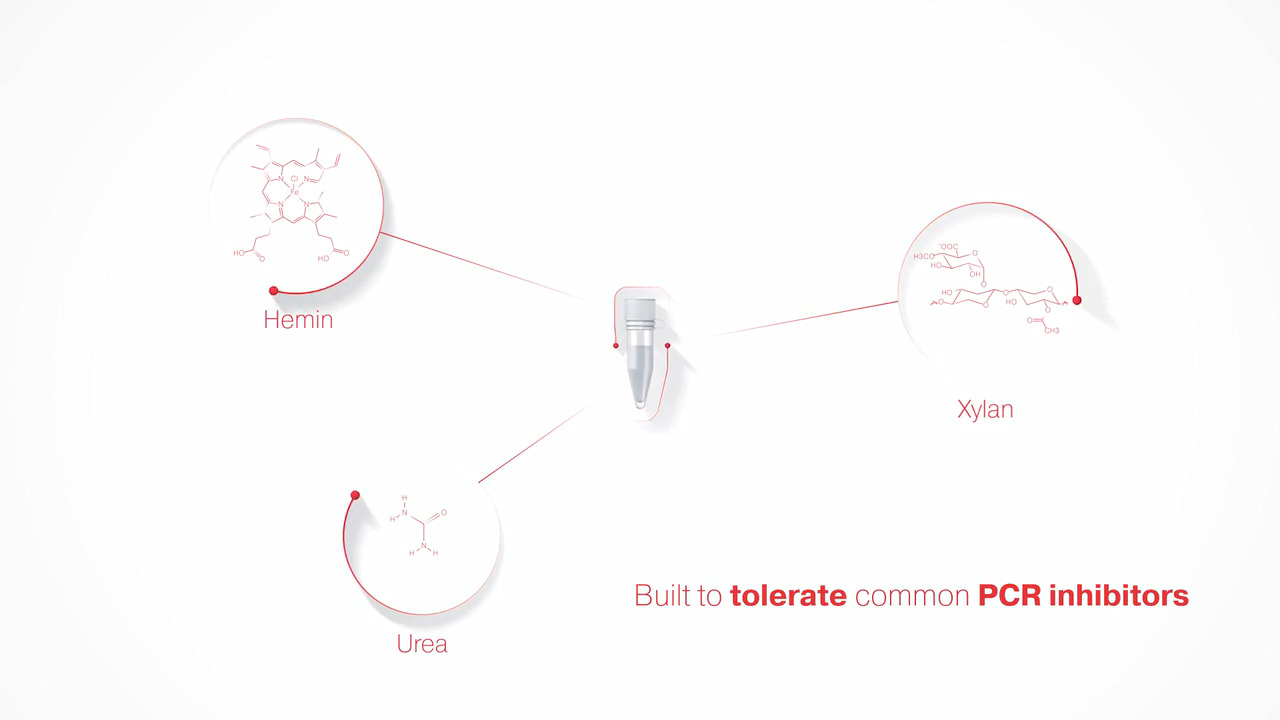
Platinum II Taq Hot-Start DNA Polymerase exhibits tolerance to common PCR inhibitors. Watch video.
Green buffer options allow for direct gel loading and help reduce pipetting errors. DNA migration is easily tracked with two dyes that are readily visible during electrophoresis.

The innovative Platinum II PCR buffer enables universal primer annealing protocol by isostabilizing primer-template duplex structures. When amplifying with Platinum II Taq DNA Polymerase, a single 60°C annealing temperature can be used for any primer pair.

Watch the benefits of universal annealing feature.
Platinum II Taq Hot-Start DNA Polymerase is four times faster than conventional Taq DNA polymerase. Watch video.
Platinum II Taq Hot-Start DNA Polymerase exhibits tolerance to common PCR inhibitors. Watch video.
Green buffer options allow for direct gel loading and help reduce pipetting errors. DNA migration is easily tracked with two dyes that are readily visible during electrophoresis.

Platinum Taq DNA polymerases: Selection guide
Platinum II Taq Hot-Start DNA Polymerase |
Platinum Taq DNA Polymerase | |
|---|---|---|
| Universal annealing protocol | Yes | No |
| Speed | 15 sec/kb | 1 min/kb |
| Flexible extension step* | Yes | No |
| Inhibitor tolerance | Yes | No |
| Target length | Up to 5 kb | Up to 5 kb |
| Hot-start modification | Antibody-mediated | Antibody-mediated |
| Fidelity versus Taq DNA Polymerase | 1x | 1x |
| Amplicon overhangs | 3’A | 3’A |
| Benchtop stability of assembled PCR reactions | 24 h | 24 h |
| GC-rich amplification | Yes | Yes |
| Certified low level of residual human and bacterial DNA‡ | Yes (≤1 copy of bacterial | No |
| Master mix formats | Colorless Green** | Colorless Green** |
| Stand-alone enzyme formats | Colorless† | Colorless Green** |
*The extension step can be extended up to 60 sec/kb without the effect on specificity.
**Direct gel loading with green buffer options.
†Green buffer available as separate item for use with stand-alone enzyme for direct loading gel.
‡During manufacturing of Platinum II Taq Hot-Start DNA Polymerase, strict measures are taken to control and verify by qPCR that no more than one copy of residual bacterial genomic DNA is present per unit of the polymerase.
Benchmarking data on Platinum II Taq Hot-Start DNA Polymerase
Engineered Platinum II Taq Hot-Start DNA Polymerase enables fast cycling of assays in as few as 30 minutes. The universal primer annealing temperature enables cycling of shorter and longer amplicons together using the same protocol.

Figure 3. Time saving enabled by assay co-cycling. PCR assays using conventional PCR reagents require specific protocols for amplification of each DNA fragment because of the different primer annealing temperatures and extension steps. Therefore, with traditional PCR reagents, multiple targets often cannot be amplified together in the same PCR run. With Platinum II Taq Hot-Start DNA Polymerase, different PCR assays can be cycled together using one protocol with a universal primer annealing temperature and the extension step selected for the longest fragment to be amplified. Moreover, Platinum II Taq Hot-Start DNA Polymerase is a fast DNA polymerase, delivering PCR results in as little as 30 minutes.

Figure 4. Platinum II Taq Hot-Start DNA Polymerase enables cycling of shorter and longer amplicons together. 132 bp, 251 bp, 1,005 bp, and 3.9 kb fragments were amplified from 50 ng of human genomic DNA in 50 μL reactions using Platinum II Taq Hot-Start DNA Polymerase or other hot-start DNA polymerases: (A) NEB OneTaq™ Hot Start DNA Polymerase, (B) Qiagen Fast Cycling™ PCR Kit, (C) Roche FastStart™ Taq DNA Polymerase. The same protocol was used for all four targets with the annealing and extension settings indicated. The size marker used is Thermo Scientific ZipRuler Express DNA Ladder 2.
The higher synthesis rate of Platinum II Taq Hot-Start DNA Polymerase allows 2 times faster PCR results compared to other hot-start Taq DNA polymerases.

Figure 5. Fast cycling reduces PCR run time. Amplification of a 529 bp fragment from 50 ng of human genomic DNA in 50 μL reactions for 35 cycles was carried out using Platinum II Taq Hot-Start DNA Polymerase and hot-start DNA polymerases from other suppliers: (A) Sigma-Aldrich KAPA2G™ Fast HotStart PCR Kit, (B) NEB OneTaq™ Hot Start DNA Polymerase, (C) Promega GoTaq™ G2 DNA Polymerase, (D) Toyobo Quick Taq™ HS DyeMix, (E) Roche FastStart™ Taq DNA Polymerase, and (F) Sigma-Aldrich JumpStart™ Taq DNA Polymerase. Cycling times for each polymerase are shown in purple, while ramping times on the ProFlex PCR System (6°C/sec peak block ramp rate) are shown in red. PCR product analysis in 1% TAE agarose gels is presented below the graph. The size marker used is the ZipRuler Express DNA Ladder 2.
The sensitivity of Platinum II Taq Hot-Start DNA Polymerase enables successful amplification of specific product in experiments where there is a limited amount of starting material, or the target DNA is in low concentration. .

Figure 6. High sensitivity and reliable amplification from low amounts of input DNA. Amplification of a 529 bp fragment from 0 (no template control); 0.016; 0.08; 0.4; 2; 10; 50; 250 ng of human genomic DNA were amplified in 50 μL PCR reactions using Platinum II Taq Hot-Start DNA Polymerase or competitor DNA polymerases (A: KAPA2G™ Fast HotStart, B: NEB OneTaq™ Hot Start, C: Promega GoTaq™ G2, D: Sigma JumpStart™ Taq, and E: Takara™ Taq HS Perfect Mix). The estimated copy number is ~5 copies per 0.016 ng of human genomic DNA. The molecular weight marker is ZipRuler Express DNA Ladder 2.
Platinum II Taq Hot-Start DNA Polymerase exhibits resistance to inhibitors and helps enable successful amplification of samples with suboptimal purity.

Figure 7. Resistance to inhibitors. Amplification of a 1 kb fragment from human genomic DNA using Platinum II Taq Hot-Start DNA Polymerase or competitor DNA polymerases (A: KAPA2G™ Robust HotStart, B: NEB OneTaq™ Hot Start, C: Promega GoTaq™ G2, and D: Takara™ Taq Hot Start Version) in reaction mixtures containing: 1: humic acid (up to final concentration of 1.3 µg/mL), 2: hemin (up to final concentration of 6 µM), 3: xylan (up to final concentration of 0.26 mg/mL), or 4: no inhibitor control. The molecular weight marker used is ZipRuler Express DNA Ladder 2.

Figure 8. Amplification of DNA extracted from FFPE tissue samples. Amplification of a 527 bp fragment from varying amounts of DNA extracted from mouse FFPE tissue samples using Platinum II Taq Hot-Start DNA polymerase. RecoverAll Total Nucleic Acid Isolation Kit for FFPE was used for DNA extraction. NTC: no template control. PC: positive control from 1 ng of purified mouse genomic DNA. The molecular weight marker used is ZipRuler Express DNA Ladder 2.
The formulation of Platinum II Taq Hot-Start DNA Polymerase and 2X Master Mixes allows for amplification of versatile range of targets, from AT-rich to GC-rich. A separate vial of Platinum GC Enhancer is provided for specific amplification and improved yields of targets with high-GC content.

PCR fragments generated by Platinum II Taq Hot-Start DNA Polymerase work well for Sanger sequencing. The enzyme’s superior performance, universal primer annealing, and fast synthesis enable generation of PCR amplicons for Sanger sequencing, with ease and simplicity.

Figure 10. High-quality Sanger sequencing results. A 1.6 kb PCR fragment amplified by Platinum II Taq Hot-Start DNA Polymerase was Sanger sequenced using Applied Biosystems 3130xl Genetic Analyzer. Data reported by the KB basecaller of the built-in sequencing analysis software is shown. Clear range read length (CRL) is defined as the longest uninterrupted segment of bases at a given Quality Value (QV). QV20 corresponds to 1% probability of a base call error and QV30 corresponds to 0.1%. QV>20 is considered high quality and acceptable in most cases.
Engineered Platinum II Taq Hot-Start DNA Polymerase enables fast cycling of assays in as few as 30 minutes. The universal primer annealing temperature enables cycling of shorter and longer amplicons together using the same protocol.

Figure 3. Time saving enabled by assay co-cycling. PCR assays using conventional PCR reagents require specific protocols for amplification of each DNA fragment because of the different primer annealing temperatures and extension steps. Therefore, with traditional PCR reagents, multiple targets often cannot be amplified together in the same PCR run. With Platinum II Taq Hot-Start DNA Polymerase, different PCR assays can be cycled together using one protocol with a universal primer annealing temperature and the extension step selected for the longest fragment to be amplified. Moreover, Platinum II Taq Hot-Start DNA Polymerase is a fast DNA polymerase, delivering PCR results in as little as 30 minutes.

Figure 4. Platinum II Taq Hot-Start DNA Polymerase enables cycling of shorter and longer amplicons together. 132 bp, 251 bp, 1,005 bp, and 3.9 kb fragments were amplified from 50 ng of human genomic DNA in 50 μL reactions using Platinum II Taq Hot-Start DNA Polymerase or other hot-start DNA polymerases: (A) NEB OneTaq™ Hot Start DNA Polymerase, (B) Qiagen Fast Cycling™ PCR Kit, (C) Roche FastStart™ Taq DNA Polymerase. The same protocol was used for all four targets with the annealing and extension settings indicated. The size marker used is Thermo Scientific ZipRuler Express DNA Ladder 2.
The higher synthesis rate of Platinum II Taq Hot-Start DNA Polymerase allows 2 times faster PCR results compared to other hot-start Taq DNA polymerases.

Figure 5. Fast cycling reduces PCR run time. Amplification of a 529 bp fragment from 50 ng of human genomic DNA in 50 μL reactions for 35 cycles was carried out using Platinum II Taq Hot-Start DNA Polymerase and hot-start DNA polymerases from other suppliers: (A) Sigma-Aldrich KAPA2G™ Fast HotStart PCR Kit, (B) NEB OneTaq™ Hot Start DNA Polymerase, (C) Promega GoTaq™ G2 DNA Polymerase, (D) Toyobo Quick Taq™ HS DyeMix, (E) Roche FastStart™ Taq DNA Polymerase, and (F) Sigma-Aldrich JumpStart™ Taq DNA Polymerase. Cycling times for each polymerase are shown in purple, while ramping times on the ProFlex PCR System (6°C/sec peak block ramp rate) are shown in red. PCR product analysis in 1% TAE agarose gels is presented below the graph. The size marker used is the ZipRuler Express DNA Ladder 2.
The sensitivity of Platinum II Taq Hot-Start DNA Polymerase enables successful amplification of specific product in experiments where there is a limited amount of starting material, or the target DNA is in low concentration. .

Figure 6. High sensitivity and reliable amplification from low amounts of input DNA. Amplification of a 529 bp fragment from 0 (no template control); 0.016; 0.08; 0.4; 2; 10; 50; 250 ng of human genomic DNA were amplified in 50 μL PCR reactions using Platinum II Taq Hot-Start DNA Polymerase or competitor DNA polymerases (A: KAPA2G™ Fast HotStart, B: NEB OneTaq™ Hot Start, C: Promega GoTaq™ G2, D: Sigma JumpStart™ Taq, and E: Takara™ Taq HS Perfect Mix). The estimated copy number is ~5 copies per 0.016 ng of human genomic DNA. The molecular weight marker is ZipRuler Express DNA Ladder 2.
Platinum II Taq Hot-Start DNA Polymerase exhibits resistance to inhibitors and helps enable successful amplification of samples with suboptimal purity.

Figure 7. Resistance to inhibitors. Amplification of a 1 kb fragment from human genomic DNA using Platinum II Taq Hot-Start DNA Polymerase or competitor DNA polymerases (A: KAPA2G™ Robust HotStart, B: NEB OneTaq™ Hot Start, C: Promega GoTaq™ G2, and D: Takara™ Taq Hot Start Version) in reaction mixtures containing: 1: humic acid (up to final concentration of 1.3 µg/mL), 2: hemin (up to final concentration of 6 µM), 3: xylan (up to final concentration of 0.26 mg/mL), or 4: no inhibitor control. The molecular weight marker used is ZipRuler Express DNA Ladder 2.

Figure 8. Amplification of DNA extracted from FFPE tissue samples. Amplification of a 527 bp fragment from varying amounts of DNA extracted from mouse FFPE tissue samples using Platinum II Taq Hot-Start DNA polymerase. RecoverAll Total Nucleic Acid Isolation Kit for FFPE was used for DNA extraction. NTC: no template control. PC: positive control from 1 ng of purified mouse genomic DNA. The molecular weight marker used is ZipRuler Express DNA Ladder 2.
The formulation of Platinum II Taq Hot-Start DNA Polymerase and 2X Master Mixes allows for amplification of versatile range of targets, from AT-rich to GC-rich. A separate vial of Platinum GC Enhancer is provided for specific amplification and improved yields of targets with high-GC content.

PCR fragments generated by Platinum II Taq Hot-Start DNA Polymerase work well for Sanger sequencing. The enzyme’s superior performance, universal primer annealing, and fast synthesis enable generation of PCR amplicons for Sanger sequencing, with ease and simplicity.

Figure 10. High-quality Sanger sequencing results. A 1.6 kb PCR fragment amplified by Platinum II Taq Hot-Start DNA Polymerase was Sanger sequenced using Applied Biosystems 3130xl Genetic Analyzer. Data reported by the KB basecaller of the built-in sequencing analysis software is shown. Clear range read length (CRL) is defined as the longest uninterrupted segment of bases at a given Quality Value (QV). QV20 corresponds to 1% probability of a base call error and QV30 corresponds to 0.1%. QV>20 is considered high quality and acceptable in most cases.
Platinum II Taq Hot-Start DNA Polymerase: Cited and quoted
Microbiota references
Cancer research references
Developmental biology references
| Usage | Publications |
|---|---|
| Two-step RT-PCR with human and mouse pluripotent stem cells | Esseltine JL, Brooks CR, Edwards NA et al. (2020) Dynamic regulation of connexins in stem cell pluripotency. Stem Cells 38:52–66. |
| MicroRNA-binding site cloning of bovine DNA | Shen X, Tang J, Ru W et al. (2021) CircINSR regulates fetal bovine muscle and fat development. Front Cell Dev Bio 8:615638. |
Disease models references
Gene regulation references
| Usage | Publications |
|---|---|
| GC-rich PCR of sea lamprey gene, followed by cloning and sequencing | Gong N, Ferreira-Martins D, McCormick SD et al. (2020) Divergent genes encoding the putative receptors for growth hormone and prolactin in sea lamprey display distinct patterns of expression. Sci Rep 10(1):1674. |
| Two-step RT-PCR with cDNA from goat primary cells for gene expression analysis | Luan Z, Fan X, Song H et al. (2019) Testosterone promotes GPX5 expression of goat epididymal epithelial cells cultured in vitro. In Vitro Cell Dev Biol Anim 55(9):677–685. |
| Chromatin immunoprecipitation (ChIP) assays | Sun D. (2019) Chromatin Immunoprecipitation Assay to Analyze the Effect of G-Quadruplex Interactive Agents on the Binding of RNA Polymerase II and Transcription Factors to a Target Promoter Region. Methods Mol Biol 2035:233–242. |
Phylogenomics references
| Usage | Publications |
|---|---|
| GC-rich PCR of flagellate DNA | Hackl T, Martin R, Barenhoff K et al. (2020) Four high-quality draft genome assemblies of the marine heterotrophic nanoflagellate Cafeteria roenbergensis. Sci Data 7(1):29. |
| Subcloning of E. coli genomic DNA fragment | Prahlad J, Yuan Y, Lin J et al. (2020) The DUF328 family member YaaA is a DNA-binding protein with a novel fold. J Biol Chem 295(41):14236–14247. |
| PCR of mitochondrial DNA from blood samples | Wharton D, Morey KC, Hanner R. (2021) Maternal inheritance of mitochondrial DNA in mice after inter-species hybridization and 138 generations of backcrossing. Mitochondrial DNA A DNA Mapp Seq Anal 32(2):73–75. |
Plant biology references
Viral research references
| Usage | Publications |
|---|---|
| PCR with DNA from FFPE samples for detection of viral (EBV, HPV) sequences | Gupta I, Al Farsi H, Jabeen A et al. (2020) High-Risk Human Papillomaviruses and Epstein-Barr Virus in Colorectal Cancer and Their Association with Clinicopathological Status. Pathogens 9(6):452. |
| Amplification of fowl adenoviral DNA from allantoic samples, followed by Sanger sequencing | Wibowo MH, Sahesty A, Mahardika BK et al. (2019) Epizootiology, Clinical Signs, and Phylogenetic Analysis of Fowl Adenovirus in Chicken Farms in Indonesia from 2018 to 2019. Avian Dis 63(4):619–624. |
Microbiota references
Cancer research references
Developmental biology references
| Usage | Publications |
|---|---|
| Two-step RT-PCR with human and mouse pluripotent stem cells | Esseltine JL, Brooks CR, Edwards NA et al. (2020) Dynamic regulation of connexins in stem cell pluripotency. Stem Cells 38:52–66. |
| MicroRNA-binding site cloning of bovine DNA | Shen X, Tang J, Ru W et al. (2021) CircINSR regulates fetal bovine muscle and fat development. Front Cell Dev Bio 8:615638. |
Disease models references
Gene regulation references
| Usage | Publications |
|---|---|
| GC-rich PCR of sea lamprey gene, followed by cloning and sequencing | Gong N, Ferreira-Martins D, McCormick SD et al. (2020) Divergent genes encoding the putative receptors for growth hormone and prolactin in sea lamprey display distinct patterns of expression. Sci Rep 10(1):1674. |
| Two-step RT-PCR with cDNA from goat primary cells for gene expression analysis | Luan Z, Fan X, Song H et al. (2019) Testosterone promotes GPX5 expression of goat epididymal epithelial cells cultured in vitro. In Vitro Cell Dev Biol Anim 55(9):677–685. |
| Chromatin immunoprecipitation (ChIP) assays | Sun D. (2019) Chromatin Immunoprecipitation Assay to Analyze the Effect of G-Quadruplex Interactive Agents on the Binding of RNA Polymerase II and Transcription Factors to a Target Promoter Region. Methods Mol Biol 2035:233–242. |
Phylogenomics references
| Usage | Publications |
|---|---|
| GC-rich PCR of flagellate DNA | Hackl T, Martin R, Barenhoff K et al. (2020) Four high-quality draft genome assemblies of the marine heterotrophic nanoflagellate Cafeteria roenbergensis. Sci Data 7(1):29. |
| Subcloning of E. coli genomic DNA fragment | Prahlad J, Yuan Y, Lin J et al. (2020) The DUF328 family member YaaA is a DNA-binding protein with a novel fold. J Biol Chem 295(41):14236–14247. |
| PCR of mitochondrial DNA from blood samples | Wharton D, Morey KC, Hanner R. (2021) Maternal inheritance of mitochondrial DNA in mice after inter-species hybridization and 138 generations of backcrossing. Mitochondrial DNA A DNA Mapp Seq Anal 32(2):73–75. |
Plant biology references
Viral research references
| Usage | Publications |
|---|---|
| PCR with DNA from FFPE samples for detection of viral (EBV, HPV) sequences | Gupta I, Al Farsi H, Jabeen A et al. (2020) High-Risk Human Papillomaviruses and Epstein-Barr Virus in Colorectal Cancer and Their Association with Clinicopathological Status. Pathogens 9(6):452. |
| Amplification of fowl adenoviral DNA from allantoic samples, followed by Sanger sequencing | Wibowo MH, Sahesty A, Mahardika BK et al. (2019) Epizootiology, Clinical Signs, and Phylogenetic Analysis of Fowl Adenovirus in Chicken Farms in Indonesia from 2018 to 2019. Avian Dis 63(4):619–624. |
“We had a very troublesome genotyping primer set that gave us bands in the NTC. Then we tried the Platinum II Taq [DNA Polymerase] and it worked the first time! We also save a ton of time on the PCR for this primer set since we no longer have to do two step PCR!”
—Research Associate
Large research university, US
PCR enzymes education resources | |
| Access PCR learning resources—a knowledge base of all things PCR. | |
Molecular biology webinars | |
| View our webinars for current date to learn more about molecular biology topics. | |
PCR enzymes videos | |
| Discover product features and tips to optimize your PCR experiments. | |
Support center | |
| Find tips, troubleshooting help, and resources for your end-point PCR and cDNA applications. | |
Contact us | |
| Email or call our Technical Application Scientists for questions regarding PCR enzymes and master mixes. | |
OEM and custom PCR products | |
| Discover custom manufacturing solutions, product labeling, and packaging capabilities tailored to your specifications. | |
For Research Use Only. Not for use in diagnostic procedures.